Plant Science Research Weekly: August 26, 2022
An experimental protocol for teaching CRISPR/Cas9 in a post-graduate plant laboratory course: An analysis of mutant-edited plants without sequencing
 Experience is the best teacher, so hand-on learning is invaluable for students of biology. If you have the opportunity to teach with a laboratory course and haven’t yet incorporated a module that incorporates CRISR/Cas9, you’re in luck! Here, Mayta et al. share the design of the laboratory course they developed in which students design a knockout of a phytoene desaturase-encoding gene PDS3, which results in white plants or sectors. The module involves four tasks, each demonstrating a different aspect of gene editing (design of sgRNAs, transformation, phenotyping, and DNA extraction and analysis). As the authors observe, they used a gel-based assay to identify edited alleles, obviating the need for more expensive sequencing. Although the authors designed this course for post-graduates, I believe it could also be carried out by advanced undergraduate students. (Summary by Mary Williams @PlantTeaching) Biochem Mol Biol Educ. 10.1002/bmb.21659
Experience is the best teacher, so hand-on learning is invaluable for students of biology. If you have the opportunity to teach with a laboratory course and haven’t yet incorporated a module that incorporates CRISR/Cas9, you’re in luck! Here, Mayta et al. share the design of the laboratory course they developed in which students design a knockout of a phytoene desaturase-encoding gene PDS3, which results in white plants or sectors. The module involves four tasks, each demonstrating a different aspect of gene editing (design of sgRNAs, transformation, phenotyping, and DNA extraction and analysis). As the authors observe, they used a gel-based assay to identify edited alleles, obviating the need for more expensive sequencing. Although the authors designed this course for post-graduates, I believe it could also be carried out by advanced undergraduate students. (Summary by Mary Williams @PlantTeaching) Biochem Mol Biol Educ. 10.1002/bmb.21659
PeTriBERT : Augmenting BERT with tridimensional encoding for inverse protein folding and design
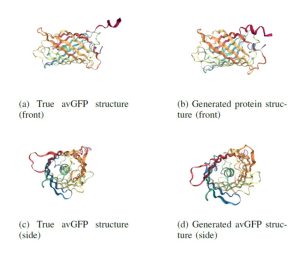 The AlphaFoldDB database recently made headlines by predicting the three-dimensional protein structure of millions of proteins based on their primary amino acid sequence. Most of these models remain to be tested, but it’s a place to start. Here, Dumortier et al. add an important complementary tool for improving our ability to correlate protein primary sequence and tertiary structure, in a tool that goes the other way; inverse protein folding. They trained their model on more than 350,000 protein sequences retrieved from the AlphaFoldDB. This model is an extension of BERT (Bidirectional Encoder Representations from Transformers), which is a machine learning / AI tool developed for language and grammar tasks (think about how Word identifies spelling and grammar mistakes in your writing). PeTriBERT (PeTriBERT: Proteins embedded in Tridimensional representation in a BERT model) enabled the researchers to generate sequences that fold to resemble GFP, of which 9/10 show no amino acid sequence homology to GFP. This tool then will enable researchers to design proteins with specific structures (and possibly similar catalytic or functional properties) but totally unique primary sequences; in silico evolution of synthetic proteins. Like so much of AI, the potential applications of this inverse-folding approach are stunning. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2022.08.10.503344
The AlphaFoldDB database recently made headlines by predicting the three-dimensional protein structure of millions of proteins based on their primary amino acid sequence. Most of these models remain to be tested, but it’s a place to start. Here, Dumortier et al. add an important complementary tool for improving our ability to correlate protein primary sequence and tertiary structure, in a tool that goes the other way; inverse protein folding. They trained their model on more than 350,000 protein sequences retrieved from the AlphaFoldDB. This model is an extension of BERT (Bidirectional Encoder Representations from Transformers), which is a machine learning / AI tool developed for language and grammar tasks (think about how Word identifies spelling and grammar mistakes in your writing). PeTriBERT (PeTriBERT: Proteins embedded in Tridimensional representation in a BERT model) enabled the researchers to generate sequences that fold to resemble GFP, of which 9/10 show no amino acid sequence homology to GFP. This tool then will enable researchers to design proteins with specific structures (and possibly similar catalytic or functional properties) but totally unique primary sequences; in silico evolution of synthetic proteins. Like so much of AI, the potential applications of this inverse-folding approach are stunning. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2022.08.10.503344
Synthetic genetic circuits as a means of reprogramming plant roots
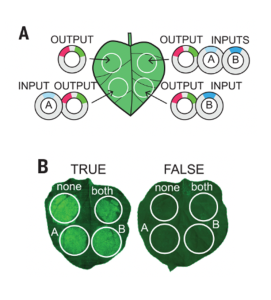 Synthetic genetic circuits offer a promising method to achieve beneficial changes in plant phenotypic traits. By combining different activator or repressor transcription factors (TFs) , the expression of target genes may be fine-tuned according to Boolean operations. Here, Brophy et al. describe a method to design and test synthetic gene circuits, and show how they can be used to modify root development in Arabidopsis thaliana. The group first designed a collection of synthetic TFs, consisting of bacterial DNA binding regions and plant promotors. They then transfected these TFs into leaves of Nicotiana benthamiana, where they were used to drive expression of green fluorescent protein, allowing rapid validation and tuning of circuit performance. Next, the researchers tested the ability of the circuits to induce tissue specific gene expression in Arabidopsis roots. Half of the tested logic gates required further tuning to achieve the expected expression patterns, demonstrating the difficulty in transferring such circuits across organisms and tissue types. Finally the group used the circuits to control expression of the mutant gene solitary root. By restricting expression of the gene circuits to lateral root stem cells, the researchers were able to selectively alter lateral root density in Arabidopsis. This ground-breaking work paves the way for further efforts to precisely modify elements of plant architecture using synthetic biology. (Summary by Rory Burke @rorby95) Science 10.1126/science.abo4326
Synthetic genetic circuits offer a promising method to achieve beneficial changes in plant phenotypic traits. By combining different activator or repressor transcription factors (TFs) , the expression of target genes may be fine-tuned according to Boolean operations. Here, Brophy et al. describe a method to design and test synthetic gene circuits, and show how they can be used to modify root development in Arabidopsis thaliana. The group first designed a collection of synthetic TFs, consisting of bacterial DNA binding regions and plant promotors. They then transfected these TFs into leaves of Nicotiana benthamiana, where they were used to drive expression of green fluorescent protein, allowing rapid validation and tuning of circuit performance. Next, the researchers tested the ability of the circuits to induce tissue specific gene expression in Arabidopsis roots. Half of the tested logic gates required further tuning to achieve the expected expression patterns, demonstrating the difficulty in transferring such circuits across organisms and tissue types. Finally the group used the circuits to control expression of the mutant gene solitary root. By restricting expression of the gene circuits to lateral root stem cells, the researchers were able to selectively alter lateral root density in Arabidopsis. This ground-breaking work paves the way for further efforts to precisely modify elements of plant architecture using synthetic biology. (Summary by Rory Burke @rorby95) Science 10.1126/science.abo4326
PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote Arabidopsis hypocotyl cell elongation
 Responses to environmental and internal signals require the recruitment of transcription factors (TFs). TFs recognize simple DNA sequences to activate specific genes that will accomplish the required functions. DOF (DNA-binding with one finger) is a large family of plant TFs that encloses the CYCLING DOF FACTORS (CDF) subfamily. CDF are regulated by the circadian clock and also promote hypocotyl elongation and regulate abiotic stress responses., but how CDFs are recruited to specific genes is unknown. In this work, Gao et al., describe how CDF2 interacts with PHYTOCHROME-INTERACTING 4 (PIF4), a TF that promotes growth in response to light and temperature. CDF2-PIF4 interaction promotes the elongation of hypocotyl cells, co-regulating common targets such as YUC8 in cotyledons. The authors showed that the increased binding strength and specificity of CDF2 in the presence of PIF4 contributes to auxin biosynthesis through the action of YUCCA enzymes, promoting the hypocotyl growth. This article shows the importance of studying specificity within gene regulatory networks. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Plants. 10.1038/s41477-022-01213-y
Responses to environmental and internal signals require the recruitment of transcription factors (TFs). TFs recognize simple DNA sequences to activate specific genes that will accomplish the required functions. DOF (DNA-binding with one finger) is a large family of plant TFs that encloses the CYCLING DOF FACTORS (CDF) subfamily. CDF are regulated by the circadian clock and also promote hypocotyl elongation and regulate abiotic stress responses., but how CDFs are recruited to specific genes is unknown. In this work, Gao et al., describe how CDF2 interacts with PHYTOCHROME-INTERACTING 4 (PIF4), a TF that promotes growth in response to light and temperature. CDF2-PIF4 interaction promotes the elongation of hypocotyl cells, co-regulating common targets such as YUC8 in cotyledons. The authors showed that the increased binding strength and specificity of CDF2 in the presence of PIF4 contributes to auxin biosynthesis through the action of YUCCA enzymes, promoting the hypocotyl growth. This article shows the importance of studying specificity within gene regulatory networks. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Plants. 10.1038/s41477-022-01213-y
Chloroplast redox state changes mark cell-to-cell signalling in the hypersensitive response
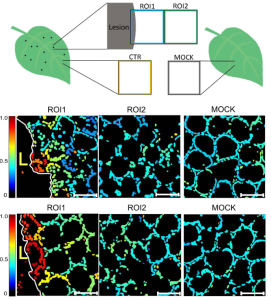 Programmed cell death (PCD) plays roles in both developmental and environmental responses across plant species. During pathogen attack, the hypersensitive response can limit spread of infection by orchestrating an organised death of cells around the infection area. A recent study by Lukan et al. demonstrated that the chloroplast redox state fluctuates depending on the proximity to the hypersensitive response death zone, confirming a role of ROS in signalling during PCD. Using spatiotemporal analysis of the redox state of potato chloroplasts in response to potato virus Y (PVY) infection, it was found that chloroplasts directly beside the death zone were highly oxidised to prevent plasmodesmata opening, thereby limiting spread of infection. Additionally, isolated mesophyll cells termed “signalling” cells, scattered close to the death zone were identified with moderately oxidized chloroplasts. The presence of these “signalling” cells was reduced in salicylic acid (SA)-deficient, infection-susceptible transgenic plants and were absent from mock-treated plants. These results indicate a role for SA in chloroplast oxidation and resistance signalling away from the death zone, in contrast with the observed SA-independent chloroplast oxidation near to the death zone and associated PCD. This study highlights the intricacies of plant cell-to-cell signalling occurring under biotic stress conditions and encourages researchers to look beyond tissue level to examine cellular molecular activities in situ. (Summary by Orla Sherwood, @orlasherwood) New Phytol. 10.1111/nph.18425
Programmed cell death (PCD) plays roles in both developmental and environmental responses across plant species. During pathogen attack, the hypersensitive response can limit spread of infection by orchestrating an organised death of cells around the infection area. A recent study by Lukan et al. demonstrated that the chloroplast redox state fluctuates depending on the proximity to the hypersensitive response death zone, confirming a role of ROS in signalling during PCD. Using spatiotemporal analysis of the redox state of potato chloroplasts in response to potato virus Y (PVY) infection, it was found that chloroplasts directly beside the death zone were highly oxidised to prevent plasmodesmata opening, thereby limiting spread of infection. Additionally, isolated mesophyll cells termed “signalling” cells, scattered close to the death zone were identified with moderately oxidized chloroplasts. The presence of these “signalling” cells was reduced in salicylic acid (SA)-deficient, infection-susceptible transgenic plants and were absent from mock-treated plants. These results indicate a role for SA in chloroplast oxidation and resistance signalling away from the death zone, in contrast with the observed SA-independent chloroplast oxidation near to the death zone and associated PCD. This study highlights the intricacies of plant cell-to-cell signalling occurring under biotic stress conditions and encourages researchers to look beyond tissue level to examine cellular molecular activities in situ. (Summary by Orla Sherwood, @orlasherwood) New Phytol. 10.1111/nph.18425
The CLP and PREP protease systems coordinate maturation and degradation of the chloroplast proteome in Arabidopsis thaliana
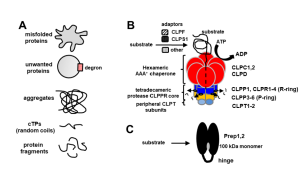 The ATP-dependent CLP chaperone-protease system is a crucial part of the peptidase network required to maintain proteostasis in the Arabidopsis chloroplast. Its substrates include misfolded and aggregated proteins of any size, and it is essential for plant growth and development. PREP1,2 peptidases are also part of this network but degrade small proteins and peptides such as cleaved chloroplast transit peptide (cTP) fragments and are targeted to both chloroplasts and mitochondria. Rowland et al. demonstrated the genetic interaction between these two systems by generating multiple combinations of higher order mutants and dissected the contributions of various components of these protease systems. The most severe growth defects were seen when both systems were defective, and the two PREP proteins are not redundant as previously thought. By using quantitative proteomics and N-terminal proteomics, the authors show that loss of CLP function causes protein folding stress in the cell and induces upregulation of a number of proteins such as an ATP-transporter, but the loss of PREP function does not impact this. Loss of both CLP and PREP1,2 function also inhibits the proper N-terminal processing of chloroplast-imported proteins by other peptidases. (Summary by Jiawen Chen @jiaaawen) New Phytol. 10.1111/nph.18426.
The ATP-dependent CLP chaperone-protease system is a crucial part of the peptidase network required to maintain proteostasis in the Arabidopsis chloroplast. Its substrates include misfolded and aggregated proteins of any size, and it is essential for plant growth and development. PREP1,2 peptidases are also part of this network but degrade small proteins and peptides such as cleaved chloroplast transit peptide (cTP) fragments and are targeted to both chloroplasts and mitochondria. Rowland et al. demonstrated the genetic interaction between these two systems by generating multiple combinations of higher order mutants and dissected the contributions of various components of these protease systems. The most severe growth defects were seen when both systems were defective, and the two PREP proteins are not redundant as previously thought. By using quantitative proteomics and N-terminal proteomics, the authors show that loss of CLP function causes protein folding stress in the cell and induces upregulation of a number of proteins such as an ATP-transporter, but the loss of PREP function does not impact this. Loss of both CLP and PREP1,2 function also inhibits the proper N-terminal processing of chloroplast-imported proteins by other peptidases. (Summary by Jiawen Chen @jiaaawen) New Phytol. 10.1111/nph.18426.
NLP7-CRF-PIN, the nitrate-cytokinin-auxin crosstalk module, conveys root nitrate signals and regulates shoot growth adaptive responses
 Nitrate, the prominent form of nitrogen used by most land plants, is a signal regulating plant growth and development. Nitrate sensing by roots not only regulates root development to facilitate nutrient foraging, but also the growth of distant plant organs. Cytokinin is a mobile signal coordinating nitrate dependent shoot growth responses. Abualia et al. show the involvement of cytokinin signalling and polar auxin transport (PAT) in nitrate-regulated shoot growth. The early response to nitrate provision coincides with changes in PAT in seedlings, and nitrate-promoted shoot growth was attenuated in the auxin efflux transporter mutants pin4 and pin7. Shoot expression of cytokinin response factors (CRF2 and CRF6) was promoted by nitrate, while the nitrate-promoted shoot growth was attenuated in crf2 and crf7 mutants. The nitrate master sensor NIN like protein 7 (NLP7) regulates the expression of PINs and CRFs in a nitrate-dependent manner. The weakened expression of cytokinin responsive ARR5 expression in nlp7 mutant roots and the promotion of root growth of nlp7 mutants by exogenous cytokinin suggests that the role of cytokinin in nitrate signalling is downstream of NLP7. This study builds on the accumulating evidence for the contribution of PAT and cytokinins in regulating nitrate-guided shoot adaptive responses by means of the NLP7-CRF-PIN module. (Summary by Lekshmy Sathee @lekshmysnair) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2122460119
Nitrate, the prominent form of nitrogen used by most land plants, is a signal regulating plant growth and development. Nitrate sensing by roots not only regulates root development to facilitate nutrient foraging, but also the growth of distant plant organs. Cytokinin is a mobile signal coordinating nitrate dependent shoot growth responses. Abualia et al. show the involvement of cytokinin signalling and polar auxin transport (PAT) in nitrate-regulated shoot growth. The early response to nitrate provision coincides with changes in PAT in seedlings, and nitrate-promoted shoot growth was attenuated in the auxin efflux transporter mutants pin4 and pin7. Shoot expression of cytokinin response factors (CRF2 and CRF6) was promoted by nitrate, while the nitrate-promoted shoot growth was attenuated in crf2 and crf7 mutants. The nitrate master sensor NIN like protein 7 (NLP7) regulates the expression of PINs and CRFs in a nitrate-dependent manner. The weakened expression of cytokinin responsive ARR5 expression in nlp7 mutant roots and the promotion of root growth of nlp7 mutants by exogenous cytokinin suggests that the role of cytokinin in nitrate signalling is downstream of NLP7. This study builds on the accumulating evidence for the contribution of PAT and cytokinins in regulating nitrate-guided shoot adaptive responses by means of the NLP7-CRF-PIN module. (Summary by Lekshmy Sathee @lekshmysnair) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2122460119
Ectopic expression of BOTRYTIS SUSCEPTIBLE1 reveals its function as a positive regulator of wound-induced cell death and plant susceptibility to Botrytis
 Programmed cell death (PCD) is a ubiquitous eukaryotic process in which specific cells are eliminated during development or in response to stress. Here, Fuqiang Cui and colleagues confirm for the first time the exact role of the BOS1 gene in the regulation of PCD in Arabidopsis thaliana. Originally identified in screen for susceptibility to the fungus Botrytis cinerea, the bos1 mutant was later shown to exhibit uncontrolled runaway cell death following wounding, suggesting BOS1 is a negative regulator of PCD. As previous bos1 T-DNA lines displayed varying transcript levels, the group first generated their own knock-out lines using CRISPR/Cas9 technology. However, none of these CRISPR/Cas9 mutants displayed the typical bos1 phenotypes in response to wounding or Botrytis infection. By resequencing the bos1 genome and generating further CRISPR/Cas9 mutants lines as well as BOS1 overexpressing plants, the group were able to confirm that bos1 is in fact a gain of function mutant, and not a loss of function mutant as originally described. The authors suggest the increase in BOS1 expression may be driven by the presence of a mannopine synthase promoter near the T-DNA insertion site, and thus urge researchers to be cautious when interpreting phenotypes in SAIL mutant lines in which the promoter is widely present. (Summary by Rory Burke @rorby95) Plant Cell 10.1093/plcell/koac206
Programmed cell death (PCD) is a ubiquitous eukaryotic process in which specific cells are eliminated during development or in response to stress. Here, Fuqiang Cui and colleagues confirm for the first time the exact role of the BOS1 gene in the regulation of PCD in Arabidopsis thaliana. Originally identified in screen for susceptibility to the fungus Botrytis cinerea, the bos1 mutant was later shown to exhibit uncontrolled runaway cell death following wounding, suggesting BOS1 is a negative regulator of PCD. As previous bos1 T-DNA lines displayed varying transcript levels, the group first generated their own knock-out lines using CRISPR/Cas9 technology. However, none of these CRISPR/Cas9 mutants displayed the typical bos1 phenotypes in response to wounding or Botrytis infection. By resequencing the bos1 genome and generating further CRISPR/Cas9 mutants lines as well as BOS1 overexpressing plants, the group were able to confirm that bos1 is in fact a gain of function mutant, and not a loss of function mutant as originally described. The authors suggest the increase in BOS1 expression may be driven by the presence of a mannopine synthase promoter near the T-DNA insertion site, and thus urge researchers to be cautious when interpreting phenotypes in SAIL mutant lines in which the promoter is widely present. (Summary by Rory Burke @rorby95) Plant Cell 10.1093/plcell/koac206



