Plant Science Research Weekly: August 16, 2023
Review. Lighting the way: Compelling open questions in photosynthesis research
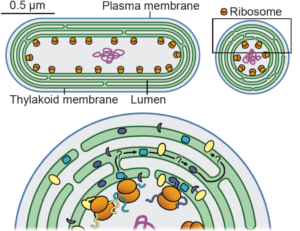 Photosynthesis is fundamental to life on Earth and a topic that all plant biologists should have a good understanding of, but it is also an incredibly complex set of processes, reactions and structures spanning great temporal and spatial distances. In this new Commentary by Eckardt et al., several experts summarize some of the fundamental unknowns being investigated in photosynthesis research. The questions focus largely on the light harvesting reactions, including the reaction center and the antenna complexes and their dynamic responses to the light and chemical environment. The authors also step beyond land plants by looking at other photosynthetic organisms, and consider unresolved questions about the evolution of photosynthesis. Open questions on stomatal regulation and the C4 pathway are also addressed. This is an engaging and compelling “must read”, and one severl in the upcoming Plant Cell Focus Issue on Photosynthesis, to be published in October, 2024. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae203
Photosynthesis is fundamental to life on Earth and a topic that all plant biologists should have a good understanding of, but it is also an incredibly complex set of processes, reactions and structures spanning great temporal and spatial distances. In this new Commentary by Eckardt et al., several experts summarize some of the fundamental unknowns being investigated in photosynthesis research. The questions focus largely on the light harvesting reactions, including the reaction center and the antenna complexes and their dynamic responses to the light and chemical environment. The authors also step beyond land plants by looking at other photosynthetic organisms, and consider unresolved questions about the evolution of photosynthesis. Open questions on stomatal regulation and the C4 pathway are also addressed. This is an engaging and compelling “must read”, and one severl in the upcoming Plant Cell Focus Issue on Photosynthesis, to be published in October, 2024. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae203
Review: Unlocking plant genetics with telomere-to-telomere genome assemblies
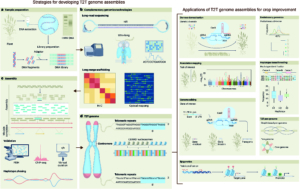 The era of complete telomere-to-telomere (T2T) plant genomes is finally here! Recent advances in long-read sequencing technologies have revolutionized genome assembly, giving rise to gapless T2T assemblies and offering unprecedented insights into genome organization and function. Why are T2T genomes so important? These assemblies provide a comprehensive, end-to-end view of a genome, covering all coding and non-coding regions. However, constructing T2T assemblies, especially for complex crop genomes with challenges like repeats, polyploidy, and heterozygosity, is a difficult task. This review by Garg et al. highlights such challenges and delves into the strategies and technological advancements necessary to overcome them. High-quality DNA extraction, improved sequencing accuracy, and the use of long-range scaffolding techniques like high-throughput chromosome conformation capture (Hi-C) and optical mapping are critical. Combining long-read sequencing and assembly algorithms, followed by manual curation, has resulted in complete T2T assemblies for crops such as rice and maize. The review also describes the current status of T2T plant genomes and their applications, including pangenomics, functional gene discovery, QTL cloning, and breeding strategies. Complete T2T assemblies promise to resolve genetic complexities in important traits like yield, disease resistance, and climate adaptation and open up new research possibilities, including developing new crop varieties for future needs. However, achieving these assemblies requires collaboration among geneticists, bioinformaticians, breeders, and other experts, along with continued investment in sequencing technologies and data integration. (Summary by Ileana Tossolini @IleanaDrt) Nature Genetics 10.1038/s41588-024-01830-7
The era of complete telomere-to-telomere (T2T) plant genomes is finally here! Recent advances in long-read sequencing technologies have revolutionized genome assembly, giving rise to gapless T2T assemblies and offering unprecedented insights into genome organization and function. Why are T2T genomes so important? These assemblies provide a comprehensive, end-to-end view of a genome, covering all coding and non-coding regions. However, constructing T2T assemblies, especially for complex crop genomes with challenges like repeats, polyploidy, and heterozygosity, is a difficult task. This review by Garg et al. highlights such challenges and delves into the strategies and technological advancements necessary to overcome them. High-quality DNA extraction, improved sequencing accuracy, and the use of long-range scaffolding techniques like high-throughput chromosome conformation capture (Hi-C) and optical mapping are critical. Combining long-read sequencing and assembly algorithms, followed by manual curation, has resulted in complete T2T assemblies for crops such as rice and maize. The review also describes the current status of T2T plant genomes and their applications, including pangenomics, functional gene discovery, QTL cloning, and breeding strategies. Complete T2T assemblies promise to resolve genetic complexities in important traits like yield, disease resistance, and climate adaptation and open up new research possibilities, including developing new crop varieties for future needs. However, achieving these assemblies requires collaboration among geneticists, bioinformaticians, breeders, and other experts, along with continued investment in sequencing technologies and data integration. (Summary by Ileana Tossolini @IleanaDrt) Nature Genetics 10.1038/s41588-024-01830-7
Control of chloroplast biogenesis by MYB-related transcription factors
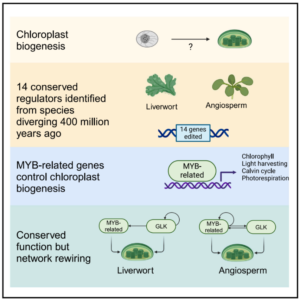 In this study, Frangedakis et al. identified MYB-related transcription factors as critical regulators of chloroplast development in land plants. Focusing on the liverwort Marchantia polymorpha and the angiosperm Arabidopsis thaliana, the team discovered that mutations in some MYB-related genes lead to pale plant phenotypes with significantly smaller chloroplasts, indicating their pivotal role in chloroplast biogenesis. Through genetic analysis and gene editing, the study revealed that these transcription factors target a broad array of genes involved in various aspects of chloroplast function, including chlorophyll biosynthesis, carbon fixation, photorespiration, and the assembly and repair of photosystems. The research also revealed that these MYB-related transcription factors work redundantly and synergistically with other known regulators, such as the GOLDEN2-LIKE (GLK) family of transcription factors, to control chloroplast development. This discovery not only reveals the regulatory mechanisms governing chloroplast development but also opens new avenues for enhancing crop productivity by manipulating these master regulators. (Summary by Amarachi Ezeoke) Cell 10.1016/j.cell.2024.06.039
In this study, Frangedakis et al. identified MYB-related transcription factors as critical regulators of chloroplast development in land plants. Focusing on the liverwort Marchantia polymorpha and the angiosperm Arabidopsis thaliana, the team discovered that mutations in some MYB-related genes lead to pale plant phenotypes with significantly smaller chloroplasts, indicating their pivotal role in chloroplast biogenesis. Through genetic analysis and gene editing, the study revealed that these transcription factors target a broad array of genes involved in various aspects of chloroplast function, including chlorophyll biosynthesis, carbon fixation, photorespiration, and the assembly and repair of photosystems. The research also revealed that these MYB-related transcription factors work redundantly and synergistically with other known regulators, such as the GOLDEN2-LIKE (GLK) family of transcription factors, to control chloroplast development. This discovery not only reveals the regulatory mechanisms governing chloroplast development but also opens new avenues for enhancing crop productivity by manipulating these master regulators. (Summary by Amarachi Ezeoke) Cell 10.1016/j.cell.2024.06.039
Plant rheostat BAP2 determines the direction of ER stress tolerance mechanisms
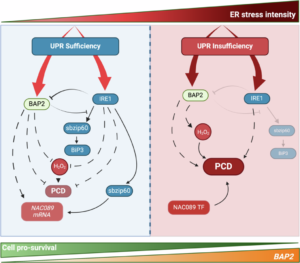 When protein folding is disrupted by abiotic or biotic stresses, cells can experience ER stress. Sensors like inositol-requiring enzyme 1 (IRE1) detect this stress, triggering the unfolded protein response (UPR) pathway. UPR signaling activates genes that restore proteostasis and maintain cellular vitality. However, under chronic ER stress, a pro-death strategy is initiated, leading to programmed cell death. But which route is taken and how this is controlled at the molecular level has been a long-standing question in ER stress research. Recently, Pastor-Cantizano and colleagues made a significant discovery in this area. They utilized the natural genetic variation in Arabidopsis to perform ER stress sensitivity assays, coupled with quantitative trait locus analyses, identifying BON-ASSOCIATED PROTEIN2 (BAP2) as a key player. Their analysis of bap2 mutants, combined with other UPR pathway components, revealed that BAP2 directs the UPR away from pro-death strategies when the stress is manageable. Conversely, under chronic stress, when the UPR can no longer sustain a pro-life response, BAP2 activates programmed cell death. This novel function of BAP2 as a rheostat, acting as a decision-maker depending on stress strength and duration, is just the beginning of unraveling the regulatory mechanisms controlling ER stress in plants. (Summary by Thomas Depaepe @thdpaepe). Nature Comms 10.1038/s41467-024-50105-6
When protein folding is disrupted by abiotic or biotic stresses, cells can experience ER stress. Sensors like inositol-requiring enzyme 1 (IRE1) detect this stress, triggering the unfolded protein response (UPR) pathway. UPR signaling activates genes that restore proteostasis and maintain cellular vitality. However, under chronic ER stress, a pro-death strategy is initiated, leading to programmed cell death. But which route is taken and how this is controlled at the molecular level has been a long-standing question in ER stress research. Recently, Pastor-Cantizano and colleagues made a significant discovery in this area. They utilized the natural genetic variation in Arabidopsis to perform ER stress sensitivity assays, coupled with quantitative trait locus analyses, identifying BON-ASSOCIATED PROTEIN2 (BAP2) as a key player. Their analysis of bap2 mutants, combined with other UPR pathway components, revealed that BAP2 directs the UPR away from pro-death strategies when the stress is manageable. Conversely, under chronic stress, when the UPR can no longer sustain a pro-life response, BAP2 activates programmed cell death. This novel function of BAP2 as a rheostat, acting as a decision-maker depending on stress strength and duration, is just the beginning of unraveling the regulatory mechanisms controlling ER stress in plants. (Summary by Thomas Depaepe @thdpaepe). Nature Comms 10.1038/s41467-024-50105-6
SAM-seq: A novel approach for unraveling plant epigenetic complexity
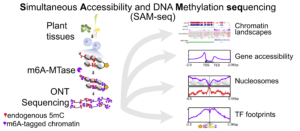 The complex interplay between nucleosome positioning, DNA methylation, and chromatin accessibility is crucial for genome regulation in eukaryotic organisms. However, current chromatin profiling methods, which rely on short-read sequencing, fail to characterize highly repetitive genomic regions and cannot detect multiple chromatin features simultaneously. In a recent study, Leduque et al. addressed these challenges by introducing a novel approach called SAM-seq (Simultaneous Accessibility and DNA Methylation sequencing). They adapted the m6A-tagged chromatin accessibility assay, SMAC-seq, for use in purified plant nuclei and employed long-read nanopore sequencing technology to assess both chromatin accessibility and DNA methylation simultaneously. SAM-seq proved effective not only in the model organism Arabidopsis but also in a complex plant genome such as maize. SAM-seq allowed for a detailed examination of accessibility and DNA methylation, particularly in subnucleosomal regions associated with genes, transposable elements, and centromeric repeats. Notably, the power and sensitivity of SAM-seq facilitated the detection of short-scale changes in accessibility and the identification of cis-regulatory regions occupied by transcription factors in vivo. Additionally, the study revealed cellular heterogeneity within chromatin domains with opposing chromatin marks, suggesting that bivalency reflects cell-specific regulatory mechanisms rather than a uniform chromatin state. SAM-seq also offers an all-in-one tool to generate de novo genome assemblies together with detailed epigenome information. Its technical simplicity, reproducibility, scalability, and cost-effectiveness make it a valuable tool for studying chromatin regulation in both model and non-model plant species. (Summary by Ileana Tossolini @IleanaDrt) Nucleic Acids Res. 10.1093/nar/gkae306
The complex interplay between nucleosome positioning, DNA methylation, and chromatin accessibility is crucial for genome regulation in eukaryotic organisms. However, current chromatin profiling methods, which rely on short-read sequencing, fail to characterize highly repetitive genomic regions and cannot detect multiple chromatin features simultaneously. In a recent study, Leduque et al. addressed these challenges by introducing a novel approach called SAM-seq (Simultaneous Accessibility and DNA Methylation sequencing). They adapted the m6A-tagged chromatin accessibility assay, SMAC-seq, for use in purified plant nuclei and employed long-read nanopore sequencing technology to assess both chromatin accessibility and DNA methylation simultaneously. SAM-seq proved effective not only in the model organism Arabidopsis but also in a complex plant genome such as maize. SAM-seq allowed for a detailed examination of accessibility and DNA methylation, particularly in subnucleosomal regions associated with genes, transposable elements, and centromeric repeats. Notably, the power and sensitivity of SAM-seq facilitated the detection of short-scale changes in accessibility and the identification of cis-regulatory regions occupied by transcription factors in vivo. Additionally, the study revealed cellular heterogeneity within chromatin domains with opposing chromatin marks, suggesting that bivalency reflects cell-specific regulatory mechanisms rather than a uniform chromatin state. SAM-seq also offers an all-in-one tool to generate de novo genome assemblies together with detailed epigenome information. Its technical simplicity, reproducibility, scalability, and cost-effectiveness make it a valuable tool for studying chromatin regulation in both model and non-model plant species. (Summary by Ileana Tossolini @IleanaDrt) Nucleic Acids Res. 10.1093/nar/gkae306
Altering cold-regulated gene expression decouples the salicylic acid–growth trade-off in Arabidopsis
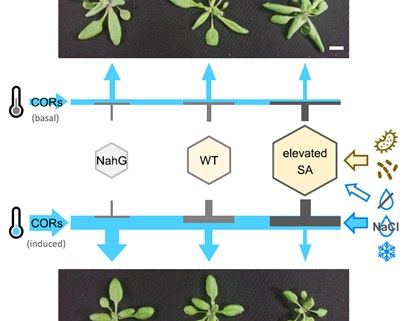
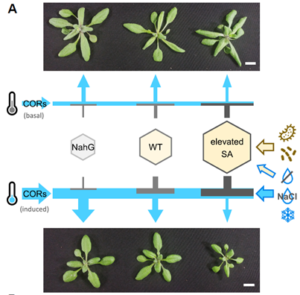 Enhancing plant defense against pathogens and pests often incurs a cost to essential physiological functions such as growth and reproduction, creating a trade-off. This phenomenon is evident in genetic studies across various plant models and agricultural breeding programs. Salicylic acid (SA) is a well-known phytohormone essential for basal immunity and systemic acquired resistance; however, its overaccumulation penalizes growth. Ortega and colleagues established a panel of Arabidopsis transgenic lines expressing a broad range of SA levels to study the effects of SA on growth, disease resistance, and abiotic stress tolerance. Interestingly, they observed that cold-regulated (COR) genes are repressed by increasing SA levels in a dose-dependent manner. Consequently, high-SA (hiSA) lines, which are smaller under unstressed conditions, grow even less when the temperature is lowered. However, strikingly, if COR genes are constitutively overexpressed in the hiSA lines, the growth defect can be rescued without compromising defense. This breakthrough suggests that the low-temperature response in plant can decouple the SA-mediated growth-defense trade-off, which potentially serving as an important strategy for balancing plant growth and immunity, thereby improving agricultural productivity. (Summary by Ching Chan @ntnuchanlab) Plant Cell 10.1093/plcell/koae210
Enhancing plant defense against pathogens and pests often incurs a cost to essential physiological functions such as growth and reproduction, creating a trade-off. This phenomenon is evident in genetic studies across various plant models and agricultural breeding programs. Salicylic acid (SA) is a well-known phytohormone essential for basal immunity and systemic acquired resistance; however, its overaccumulation penalizes growth. Ortega and colleagues established a panel of Arabidopsis transgenic lines expressing a broad range of SA levels to study the effects of SA on growth, disease resistance, and abiotic stress tolerance. Interestingly, they observed that cold-regulated (COR) genes are repressed by increasing SA levels in a dose-dependent manner. Consequently, high-SA (hiSA) lines, which are smaller under unstressed conditions, grow even less when the temperature is lowered. However, strikingly, if COR genes are constitutively overexpressed in the hiSA lines, the growth defect can be rescued without compromising defense. This breakthrough suggests that the low-temperature response in plant can decouple the SA-mediated growth-defense trade-off, which potentially serving as an important strategy for balancing plant growth and immunity, thereby improving agricultural productivity. (Summary by Ching Chan @ntnuchanlab) Plant Cell 10.1093/plcell/koae210
Development of a structure-switching aptamer-based nanosensor for salicylic
acid detection
 Aptamers are single-stranded DNAs that fold into tertiary structures that bind specifically to various targets, allowing them to serve as probes or sensors. In this study, Chen et al. introduce a cutting-edge aptamer-based nanopore thin film sensor designed to detect salicylic acid (SA), an important compound involved in plant immunity and human health. Traditional methods like high-performance liquid chromatography (HPLC) and mass spectrometry face limitations due to SA’s small size and lack of reactive groups. The new sensor addresses these challenges by offering high sensitivity and specificity. A key innovation in this study is the use of structure-switching SELEX, a technique that immobilizes the aptamer library rather than the target molecule. This approach enabled the identification of a highly specific SA aptamer that effectively distinguishes SA from its metabolites and structural analogs. The sensor demonstrated a detection limit of 0.1 μM SA and showed excellent performance in both buffer solutions and plant extracts from Arabidopsis and rice. Additionally, the sensor’s binding kinetics were optimized, achieving equilibrium within 5 minutes for low SA concentrations. This rapid and cost-effective sensor surpasses traditional antibody-based methods and offers a robust tool for on-site SA detection. Its high sensitivity and specificity make it a valuable asset for plant science research and potentially for broader applications in life sciences and medicine. (Summary by Amarachi Ezeoke) Biosens Bioelectron 10.1016/j.bios.2019.111342
Aptamers are single-stranded DNAs that fold into tertiary structures that bind specifically to various targets, allowing them to serve as probes or sensors. In this study, Chen et al. introduce a cutting-edge aptamer-based nanopore thin film sensor designed to detect salicylic acid (SA), an important compound involved in plant immunity and human health. Traditional methods like high-performance liquid chromatography (HPLC) and mass spectrometry face limitations due to SA’s small size and lack of reactive groups. The new sensor addresses these challenges by offering high sensitivity and specificity. A key innovation in this study is the use of structure-switching SELEX, a technique that immobilizes the aptamer library rather than the target molecule. This approach enabled the identification of a highly specific SA aptamer that effectively distinguishes SA from its metabolites and structural analogs. The sensor demonstrated a detection limit of 0.1 μM SA and showed excellent performance in both buffer solutions and plant extracts from Arabidopsis and rice. Additionally, the sensor’s binding kinetics were optimized, achieving equilibrium within 5 minutes for low SA concentrations. This rapid and cost-effective sensor surpasses traditional antibody-based methods and offers a robust tool for on-site SA detection. Its high sensitivity and specificity make it a valuable asset for plant science research and potentially for broader applications in life sciences and medicine. (Summary by Amarachi Ezeoke) Biosens Bioelectron 10.1016/j.bios.2019.111342
Unveiling root trait syndromes in trees: Evolutionary insights into mycorrhizal partnerships
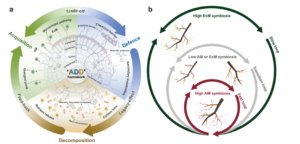 Plants, particularly long-lived trees, need to balance nutrient acquisition, herbivore and pathogen defence, and ultimately organic matter decomposition. The intricate relationship between these processes is crucial for the survival and growth of trees over their extended lifespans. Zheng et al. investigated the complex link between tree species and their mycorrhizal symbionts, focussing on how root trait syndromes evolved to optimise nutrient intake, defence, and decomposition, and leading to a proposed Acquisition-Defense-Decomposition (ADD) framework. AM (arbuscular mycorrhiza)-associated plants, which are prevalent in tropical locations, have root characteristics that promote quick nutrient acquisition, such as long root length and surface area. In contrast, EM (ecto-mycorrhiza) associated trees, which are found in temperate and boreal zones, have root features that emphasise nutrient conservation and defence, such as bigger roots and increased investment in secondary metabolites. These syndromes are also associated with decomposition processes, with AM roots contributing to quicker nutrient cycling and EM roots leading to slower decomposition rates, influencing soil carbon storage. The findings shed light on the evolutionary trade-offs that have formed root characteristics in response to environmental stressors and symbiotic interactions. By connecting root features to broader ecosystem functions, the work improves our understanding of how trees adapt to diverse ecological niches and how mycorrhizal relationships influence forest dynamics. It further emphasises the necessity of combining root trait studies and mycorrhizal ecology to guide conservation policies and forecast ecosystem responses to environmental change. (Summary by Tuyelee Das @Das_tuyelee) Nature Comms 10.1038/s41467-024-49666-3)
Plants, particularly long-lived trees, need to balance nutrient acquisition, herbivore and pathogen defence, and ultimately organic matter decomposition. The intricate relationship between these processes is crucial for the survival and growth of trees over their extended lifespans. Zheng et al. investigated the complex link between tree species and their mycorrhizal symbionts, focussing on how root trait syndromes evolved to optimise nutrient intake, defence, and decomposition, and leading to a proposed Acquisition-Defense-Decomposition (ADD) framework. AM (arbuscular mycorrhiza)-associated plants, which are prevalent in tropical locations, have root characteristics that promote quick nutrient acquisition, such as long root length and surface area. In contrast, EM (ecto-mycorrhiza) associated trees, which are found in temperate and boreal zones, have root features that emphasise nutrient conservation and defence, such as bigger roots and increased investment in secondary metabolites. These syndromes are also associated with decomposition processes, with AM roots contributing to quicker nutrient cycling and EM roots leading to slower decomposition rates, influencing soil carbon storage. The findings shed light on the evolutionary trade-offs that have formed root characteristics in response to environmental stressors and symbiotic interactions. By connecting root features to broader ecosystem functions, the work improves our understanding of how trees adapt to diverse ecological niches and how mycorrhizal relationships influence forest dynamics. It further emphasises the necessity of combining root trait studies and mycorrhizal ecology to guide conservation policies and forecast ecosystem responses to environmental change. (Summary by Tuyelee Das @Das_tuyelee) Nature Comms 10.1038/s41467-024-49666-3)
Forests endure as critical carbon sinks
 The global forest carbon sink is vital for mitigating climate change by absorbing CO2 and offsetting nearly half of fossil-fuel emissions. Over the past three decades, this sink has remained stable with minor fluctuations, but its future is threatened by deforestation, forest ageing, and climate change. Pan et al. conducted a comprehensive analysis using ground-based forest data from boreal, temperate, and tropical biomes. They found that the global forest carbon sink’s stability masks regional variations: temperate and tropical regrowth forests have increased carbon uptake, while boreal and tropical intact forests have declined. Boreal forests’ carbon sink has diminished due to intensified disturbances like wildfires and insect outbreaks. In contrast, temperate forests have increased their carbon sequestration due to extensive afforestation and reforestation, particularly in China. Tropical regrowth forests, which have expanded on previously agricultural lands, also significantly contribute to carbon sequestration. The study highlights the need for effective land management policies to limit deforestation, promote forest restoration, and improve timber-harvesting practices to protect and enhance the forest carbon sink. Ensuring the resilience of this critical climate mitigation tool requires continued investment in forest monitoring and research. (Summary by Amarachi Ezeoke) Nature 10.1038/s41586-024-07602-x
The global forest carbon sink is vital for mitigating climate change by absorbing CO2 and offsetting nearly half of fossil-fuel emissions. Over the past three decades, this sink has remained stable with minor fluctuations, but its future is threatened by deforestation, forest ageing, and climate change. Pan et al. conducted a comprehensive analysis using ground-based forest data from boreal, temperate, and tropical biomes. They found that the global forest carbon sink’s stability masks regional variations: temperate and tropical regrowth forests have increased carbon uptake, while boreal and tropical intact forests have declined. Boreal forests’ carbon sink has diminished due to intensified disturbances like wildfires and insect outbreaks. In contrast, temperate forests have increased their carbon sequestration due to extensive afforestation and reforestation, particularly in China. Tropical regrowth forests, which have expanded on previously agricultural lands, also significantly contribute to carbon sequestration. The study highlights the need for effective land management policies to limit deforestation, promote forest restoration, and improve timber-harvesting practices to protect and enhance the forest carbon sink. Ensuring the resilience of this critical climate mitigation tool requires continued investment in forest monitoring and research. (Summary by Amarachi Ezeoke) Nature 10.1038/s41586-024-07602-x