Plant Science Research Weekly: April 5th
Plant Physiology Focus issue on biotic stress
 The April 2019 issue of Plant Physiology includes a set of papers addressing biotic interactions between plant and pathogens or pests. As the editors indicate, progress on this topic has been rapid and significant. Key topics explored in reviews and research articles include: the roles of plant hormones, resistance proteins and signaling in defense, communication and nutrient exchange between plant and pathogen, and the roles of chromatin dynamics and modifications in defense. (Summary by Mary Williams) Plant Physiology. 10.1104/pp.19.00330
The April 2019 issue of Plant Physiology includes a set of papers addressing biotic interactions between plant and pathogens or pests. As the editors indicate, progress on this topic has been rapid and significant. Key topics explored in reviews and research articles include: the roles of plant hormones, resistance proteins and signaling in defense, communication and nutrient exchange between plant and pathogen, and the roles of chromatin dynamics and modifications in defense. (Summary by Mary Williams) Plant Physiology. 10.1104/pp.19.00330
Review: Tackling plant phosphate starvation by the roots
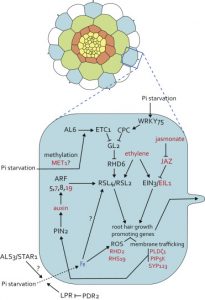 The root is remarkably responsive to nutrient status of both the local environment and the whole plant. These external and internal factors affect the expression of transporters, interactions with mycorrhizal symbionts and also the overall pattern of growth and development of the root system. In this new review, Crombez et al. review how low P leads to all of these responses. They integrate our understanding of the roles of hormones and signals into a spatial and developmental model of the root, and describe the important contributions of lateral roots and the root cap in P uptake. (Summary by Mary Williams) Devel. Cell 10.1016/j.devcel.2019.01.002
The root is remarkably responsive to nutrient status of both the local environment and the whole plant. These external and internal factors affect the expression of transporters, interactions with mycorrhizal symbionts and also the overall pattern of growth and development of the root system. In this new review, Crombez et al. review how low P leads to all of these responses. They integrate our understanding of the roles of hormones and signals into a spatial and developmental model of the root, and describe the important contributions of lateral roots and the root cap in P uptake. (Summary by Mary Williams) Devel. Cell 10.1016/j.devcel.2019.01.002
CRISPR-Chip: Detection of gene variants in unamplified DNA using graphene field-effect transistors ($)
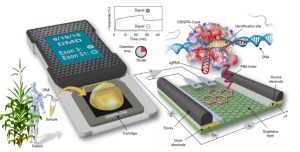 CRISPR-Chip combines the selectivity of CRISPR-Cas gene recognition with the sensitivity of a graphene-based field-effect transistor. It contains an immobilized complex of catalytically deactivated Cas9/CRISPR and an appropriate guide strand attached to a graphene surface. When this complex recognizes its target sequence, it generates an electrical signal within 15 minutes of applying the sample. What’s remarkable is that this method is extremely sensitive, and does not require DNA amplification. The authors demonstrate its ability to detect deletion mutations of the human dystrophin gene. They don’t address smaller mutants or sequence variations, but it seems likely that those improvements are on the horizon. (Summary by Mary Williams) Nature Biomed Engineering 10.1038/s41551-019-0371-x
CRISPR-Chip combines the selectivity of CRISPR-Cas gene recognition with the sensitivity of a graphene-based field-effect transistor. It contains an immobilized complex of catalytically deactivated Cas9/CRISPR and an appropriate guide strand attached to a graphene surface. When this complex recognizes its target sequence, it generates an electrical signal within 15 minutes of applying the sample. What’s remarkable is that this method is extremely sensitive, and does not require DNA amplification. The authors demonstrate its ability to detect deletion mutations of the human dystrophin gene. They don’t address smaller mutants or sequence variations, but it seems likely that those improvements are on the horizon. (Summary by Mary Williams) Nature Biomed Engineering 10.1038/s41551-019-0371-x
Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth ($)
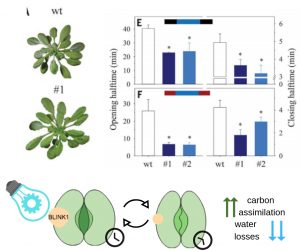 Optogenetics is a biotechnique that uses light-sensitive molecules to regulate cell activity. In plants, stomatal aperture mediates both CO2 uptake for photosynthesis and water loss by transpiration. The carbon-water trade-off control affects water use efficiency (WUE). Here, Papanatsiou et al. studied the synthetic blue light–induced K+ channel 1 (BLINK1) as an ion flux photoactivator by expressing it in the guard cells membranes of Arabidopsis. Membrane voltage kinetics showed an increase of K+ conductance in the BLINK1-guard cells in response to light. The stomatal kinetics, analyzed by gas exchange, revealed BLINK1 accelerates changes in conductance, reducing the stomatal opening and closing halftimes. Rosette area and biomass increase were documented and CO2 concentrations were not altered, meaning in an improvement of WUE without affecting carbon assimilation. Optogenetic tools are mainly known for their contributions to control mammalian neurological functions, and this study demonstrates how valuable these new strategies are to modifying plant growth responses as well. (Summary by Ana Valladares) Science. 10.1126/science.aaw0046
Optogenetics is a biotechnique that uses light-sensitive molecules to regulate cell activity. In plants, stomatal aperture mediates both CO2 uptake for photosynthesis and water loss by transpiration. The carbon-water trade-off control affects water use efficiency (WUE). Here, Papanatsiou et al. studied the synthetic blue light–induced K+ channel 1 (BLINK1) as an ion flux photoactivator by expressing it in the guard cells membranes of Arabidopsis. Membrane voltage kinetics showed an increase of K+ conductance in the BLINK1-guard cells in response to light. The stomatal kinetics, analyzed by gas exchange, revealed BLINK1 accelerates changes in conductance, reducing the stomatal opening and closing halftimes. Rosette area and biomass increase were documented and CO2 concentrations were not altered, meaning in an improvement of WUE without affecting carbon assimilation. Optogenetic tools are mainly known for their contributions to control mammalian neurological functions, and this study demonstrates how valuable these new strategies are to modifying plant growth responses as well. (Summary by Ana Valladares) Science. 10.1126/science.aaw0046
Generating novel, dual herbicide-resistant crops using CRISPR-mediated gene editing
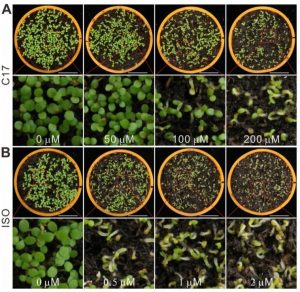 There is an ever-growing need for novel herbicides as more weeds are becoming resistant to commonly used herbicides. In a recent report from Hu et al., the authors identified that a previously described cellulose biosynthesis-inhibiting chemical, C17, possesses an herbicidal quality. The authors showed that the treatment with C17 resulted in the depletion of cellulose synthase subunits (CESA) in the plasma membrane of weeds and dicotyledonous crops while leaving monocotyledons unaffected. By combining treatment with commercially used cellulose biosynthesis inhibitors, the authors identified that C17 affects CESA through a novel mode of action. To develop new methods of herbicide treatment, the authors used CRISPR-mediated gene editing to engineer plants that were resistant to C17 by mutating the CESA3 gene. They also showed that this resistance could be coupled with resistance to other cellulose biosynthetic genes to develop a more robust, herbicide resistant crop which did not display dwarf phenotypes common in double herbicide-resistant mutants generated through traditional breeding methods. These results provide the foundation for the use of C17 as a new tool in both the generation of novel herbicide resistant crops as well as a research tool for transformation selection markers. (Summary by Nick Segerson) Plant Physiol. 10.1104/pp.18.01486
There is an ever-growing need for novel herbicides as more weeds are becoming resistant to commonly used herbicides. In a recent report from Hu et al., the authors identified that a previously described cellulose biosynthesis-inhibiting chemical, C17, possesses an herbicidal quality. The authors showed that the treatment with C17 resulted in the depletion of cellulose synthase subunits (CESA) in the plasma membrane of weeds and dicotyledonous crops while leaving monocotyledons unaffected. By combining treatment with commercially used cellulose biosynthesis inhibitors, the authors identified that C17 affects CESA through a novel mode of action. To develop new methods of herbicide treatment, the authors used CRISPR-mediated gene editing to engineer plants that were resistant to C17 by mutating the CESA3 gene. They also showed that this resistance could be coupled with resistance to other cellulose biosynthetic genes to develop a more robust, herbicide resistant crop which did not display dwarf phenotypes common in double herbicide-resistant mutants generated through traditional breeding methods. These results provide the foundation for the use of C17 as a new tool in both the generation of novel herbicide resistant crops as well as a research tool for transformation selection markers. (Summary by Nick Segerson) Plant Physiol. 10.1104/pp.18.01486
High-temporal-resolution transcriptome landscape of early maize seed development
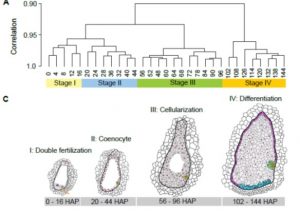 Seeds determine the yield and quality of the plants. In maize, seed development initiates from a double fertilization event where one pollen sperm fuses with the egg and the other with the central cell of female gametophyte to produce the progenitors of embryo and endosperm, respectively. The embryo carries genetic information to the next generation, and the endosperm serves as a storage tissue for seedling development. In maize, primary endosperm first undergoes several rounds of cell division without cytokinesis to form the coenocyte (a multinucleate cell), which subsequently undergoes cellularization and differentiation. Identifying the genes involved and their functions will provide insights into the genetic control of seed development and enables the improvement of seed traits. Given this, Yi et al. have constructed a high-temporal-resolution dynamic transcriptome landscape of early seed development using 31 samples collected at an interval of 4 or 6 hours within the first six days of seed development. The study revealed the genes involved in the key steps of double fertilization (4453 genes), coenocyte formation (1285), cellularization (2569) and differentiation (3614). The study also identified 1093 seed-specific genes including 110 transcription factors (TFs) among which MYB131, MYB16 and BZIP109 are critical for endosperm cellularization and MYBR81, BZIP46 and HB118 are involved in endosperm differentiation. However, further functional characterization is required to understand the precise roles of these genes and TFs in seed development. Altogether, the study provides a high-temporal-resolution atlas of gene expression that occurs in the first few days of seed development in maize that would be useful in identifying the vital players involved in determining each specific cell types of the early seed development. (Summary by Muthamilarasan Mehanathan) Plant Cell 10.1105/tpc.18.00961
Seeds determine the yield and quality of the plants. In maize, seed development initiates from a double fertilization event where one pollen sperm fuses with the egg and the other with the central cell of female gametophyte to produce the progenitors of embryo and endosperm, respectively. The embryo carries genetic information to the next generation, and the endosperm serves as a storage tissue for seedling development. In maize, primary endosperm first undergoes several rounds of cell division without cytokinesis to form the coenocyte (a multinucleate cell), which subsequently undergoes cellularization and differentiation. Identifying the genes involved and their functions will provide insights into the genetic control of seed development and enables the improvement of seed traits. Given this, Yi et al. have constructed a high-temporal-resolution dynamic transcriptome landscape of early seed development using 31 samples collected at an interval of 4 or 6 hours within the first six days of seed development. The study revealed the genes involved in the key steps of double fertilization (4453 genes), coenocyte formation (1285), cellularization (2569) and differentiation (3614). The study also identified 1093 seed-specific genes including 110 transcription factors (TFs) among which MYB131, MYB16 and BZIP109 are critical for endosperm cellularization and MYBR81, BZIP46 and HB118 are involved in endosperm differentiation. However, further functional characterization is required to understand the precise roles of these genes and TFs in seed development. Altogether, the study provides a high-temporal-resolution atlas of gene expression that occurs in the first few days of seed development in maize that would be useful in identifying the vital players involved in determining each specific cell types of the early seed development. (Summary by Muthamilarasan Mehanathan) Plant Cell 10.1105/tpc.18.00961
3D characteristics of grain reveal differences between domesticated and wild relatives in wheat and barley
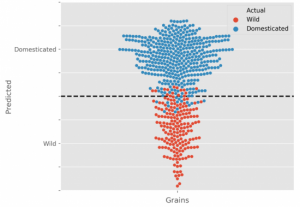 Grain related traits play key roles in determining the total grain yield. However, traditionally manual measurement can not investigate grain seed from multiple angles. To visualize a 3D structure of grain, X-ray based images were captured for a single spike/grain from multiple angles. Because of strong penetration ability, X-ray can not only capture pixels on the surface of object but also inside areas. Using this technology, a list of grain traits including grain width, length, depth, volume and surface areas were generated. Three traits of grain width, length and depth were then considered as predictors for domestication prediction; using them the R square values are between 0.95 to 0.99. Grain depth is shown to be the most critical predictor among them in the prediction model in cereals. (Summary by Zhikai Liang) Plant Journal: 10.1111/tpj.14312
Grain related traits play key roles in determining the total grain yield. However, traditionally manual measurement can not investigate grain seed from multiple angles. To visualize a 3D structure of grain, X-ray based images were captured for a single spike/grain from multiple angles. Because of strong penetration ability, X-ray can not only capture pixels on the surface of object but also inside areas. Using this technology, a list of grain traits including grain width, length, depth, volume and surface areas were generated. Three traits of grain width, length and depth were then considered as predictors for domestication prediction; using them the R square values are between 0.95 to 0.99. Grain depth is shown to be the most critical predictor among them in the prediction model in cereals. (Summary by Zhikai Liang) Plant Journal: 10.1111/tpj.14312
Single-cell RNA sequencing reveals developmental trajectories of Arabidopsis root cells
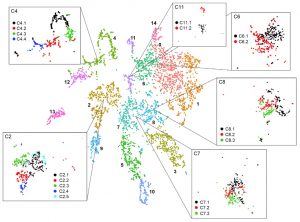 New advances in sequencing technologies have made possible the understanding of the dynamics of developmental processes in an unprecedented manner. In a recent study, Denyer, Ma, et al. profiled nearly 5,000 cells from Arabidopsis roots using single-cell RNA sequencing technology. Gene expression profiles grouped cells into clusters representing the major cell types in the root; each cluster included some well-characterized genes (i.e. PLT1, SCR, SHR), so, it was possible to assign its identity, additionally new cell-type specific genes were identified, for some of them, the predicted expression pattern was confirmed using reporter lines. Furthermore, sub-clustering allowed the identification of cells in a transitional state, so it was possible to follow a developmental trajectory. An example of these was made with the analysis of the transition from stem cells to mature trichoblast. Authors also analyzed clusters derived from cell populations of the shortroot-3 mutant, they detect the expected absence of the endodermis clusters and detected hidden defects in other cell types. This work provides an excellent spatiotemporal perspective of root cell differentiation. (Summary by Humberto Herrera-Ubaldo) Devel. Cell 10.1016/j.devcel.2019.02.022
New advances in sequencing technologies have made possible the understanding of the dynamics of developmental processes in an unprecedented manner. In a recent study, Denyer, Ma, et al. profiled nearly 5,000 cells from Arabidopsis roots using single-cell RNA sequencing technology. Gene expression profiles grouped cells into clusters representing the major cell types in the root; each cluster included some well-characterized genes (i.e. PLT1, SCR, SHR), so, it was possible to assign its identity, additionally new cell-type specific genes were identified, for some of them, the predicted expression pattern was confirmed using reporter lines. Furthermore, sub-clustering allowed the identification of cells in a transitional state, so it was possible to follow a developmental trajectory. An example of these was made with the analysis of the transition from stem cells to mature trichoblast. Authors also analyzed clusters derived from cell populations of the shortroot-3 mutant, they detect the expected absence of the endodermis clusters and detected hidden defects in other cell types. This work provides an excellent spatiotemporal perspective of root cell differentiation. (Summary by Humberto Herrera-Ubaldo) Devel. Cell 10.1016/j.devcel.2019.02.022
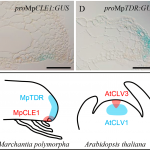
Control of proliferation in the haploid meristem by CLE peptide signaling in Marchantia polymorpha
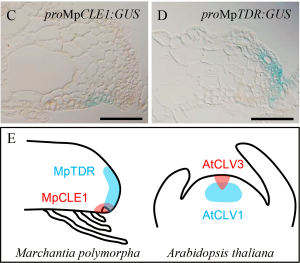 The peptide/hormones CLE (CLAVATA3/EMBRYO SURROUNDING REGION-related) and their receptors are a fascinating cell signal system that regulates meristems activity in angiosperms and is crucial for the establishment of 3D forms. How this system works in bryophytes is very relevant to understand its evolution in plants that develop different body plans. Differently to the moss Physcomitrella patens that possess a single CLE peptide similar to CLV3 type, the liverwort Marchantia polymorpha genome harbors a single copy of both known types (CLV3 and TDIF). Here, Hirakawa et al. provide an exhaustive characterization of the TDIF type peptide MpCLE1 in M. polymorpha. With a few amino acid changes, MpCLE1 can mimic the effect of the homolog AtCLE41 in Arabidopsis. Despite that, mutant and overexpressing plants of MpCLE1 and the receptor, MpTDR, suggest that they work as negative regulators of meristem activity, similar to CLV-type but the opposite function described for TDIF-type peptides in angiosperm.On the other hand, MpCLE1 loss-of-function resembles the abnormal cell division planes phenotype in TDIF gain and loss-of-function in Arabidopsis. Its function seems to be a combination of both CLE peptides types in angiosperms, showing the complex evolution pattern of this signaling system in land plants. (Summary by Facundo Romani) PLOS Genetics 10.1371/journal.pgen.1007997
The peptide/hormones CLE (CLAVATA3/EMBRYO SURROUNDING REGION-related) and their receptors are a fascinating cell signal system that regulates meristems activity in angiosperms and is crucial for the establishment of 3D forms. How this system works in bryophytes is very relevant to understand its evolution in plants that develop different body plans. Differently to the moss Physcomitrella patens that possess a single CLE peptide similar to CLV3 type, the liverwort Marchantia polymorpha genome harbors a single copy of both known types (CLV3 and TDIF). Here, Hirakawa et al. provide an exhaustive characterization of the TDIF type peptide MpCLE1 in M. polymorpha. With a few amino acid changes, MpCLE1 can mimic the effect of the homolog AtCLE41 in Arabidopsis. Despite that, mutant and overexpressing plants of MpCLE1 and the receptor, MpTDR, suggest that they work as negative regulators of meristem activity, similar to CLV-type but the opposite function described for TDIF-type peptides in angiosperm.On the other hand, MpCLE1 loss-of-function resembles the abnormal cell division planes phenotype in TDIF gain and loss-of-function in Arabidopsis. Its function seems to be a combination of both CLE peptides types in angiosperms, showing the complex evolution pattern of this signaling system in land plants. (Summary by Facundo Romani) PLOS Genetics 10.1371/journal.pgen.1007997
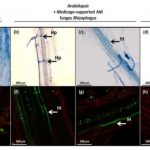
Antagonistic responses to ‘symbiotic’ arbuscular mycorrhizal fungi in the non-host Arabidopsis thaliana ($)
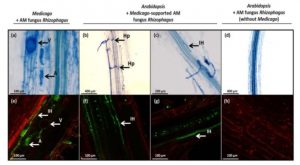 A strikingly vast array of phylogenetically distant plants are able to form intimate interactions with symbiotic arbuscular mycorrhizal (AM) fungi for the mutually beneficial exchange of nutrients. Several years of intensive research have revealed the complex regulatory networks and genetic ‘toolkits’ that enable these interactions, yet comparatively less attention is paid to understanding interactions between AM fungi and non-host plant species. Fernández et al. investigated the molecular responses of Arabidopsis thaliana during non-host interactions with the AM fungus Rhizophagus irregularis. The authors first established that Arabidopsis roots exposed to R. irregularis fungal mycelia upregulate the biosynthesis of the AM-attracting plant hormone strigolactone, similar to AM-supporting host species. This upregulation was not observed when Arabidopsis roots were exposed to beneficial or detrimental species of non-AM fungi, demonstrating that Arabidopsis maintains specific early AM-response programs that evolved to facilitate AM symbiosis. Next, the authors investigated the impact of non-host colonization in Arabidopsis roots using a host species-(Medicago truncatula)-supported AM fungal network. Microscopic analyses demonstrated that Arabidopsis supports early invasive fungal hyphopodea (entrance structures) and intraradical hyphae, although such colonization ultimately resulted in significant reductions in plant biomass. Transcriptome analysis of AM-colonized Arabidopsis roots revealed a strong upregulation of plant defenses, indicative of a robust antagonistic response. Intriguingly, leaves of AM-colonized Arabidopsis plants displayed enhanced resistance to the necrotrophic fungal pathogen Botrytis cinerea, which suggests that antagonistic non-host interactions in roots provides systemic immune benefits. Collectively, these results uncover key similarities and differences in the responses of host and non-host plant species to R. irregularis, suggestive of further complexity in the role of AM fungi in diverse plant communities. (Summary by Phil Carella) New Phytol. 10.1111/nph.15798
A strikingly vast array of phylogenetically distant plants are able to form intimate interactions with symbiotic arbuscular mycorrhizal (AM) fungi for the mutually beneficial exchange of nutrients. Several years of intensive research have revealed the complex regulatory networks and genetic ‘toolkits’ that enable these interactions, yet comparatively less attention is paid to understanding interactions between AM fungi and non-host plant species. Fernández et al. investigated the molecular responses of Arabidopsis thaliana during non-host interactions with the AM fungus Rhizophagus irregularis. The authors first established that Arabidopsis roots exposed to R. irregularis fungal mycelia upregulate the biosynthesis of the AM-attracting plant hormone strigolactone, similar to AM-supporting host species. This upregulation was not observed when Arabidopsis roots were exposed to beneficial or detrimental species of non-AM fungi, demonstrating that Arabidopsis maintains specific early AM-response programs that evolved to facilitate AM symbiosis. Next, the authors investigated the impact of non-host colonization in Arabidopsis roots using a host species-(Medicago truncatula)-supported AM fungal network. Microscopic analyses demonstrated that Arabidopsis supports early invasive fungal hyphopodea (entrance structures) and intraradical hyphae, although such colonization ultimately resulted in significant reductions in plant biomass. Transcriptome analysis of AM-colonized Arabidopsis roots revealed a strong upregulation of plant defenses, indicative of a robust antagonistic response. Intriguingly, leaves of AM-colonized Arabidopsis plants displayed enhanced resistance to the necrotrophic fungal pathogen Botrytis cinerea, which suggests that antagonistic non-host interactions in roots provides systemic immune benefits. Collectively, these results uncover key similarities and differences in the responses of host and non-host plant species to R. irregularis, suggestive of further complexity in the role of AM fungi in diverse plant communities. (Summary by Phil Carella) New Phytol. 10.1111/nph.15798