Plant Science Research Weekly: April 30th, 2021
Review: Ageing beautifully: can the benefits of seed priming be separated from a reduced lifespan trade-off?
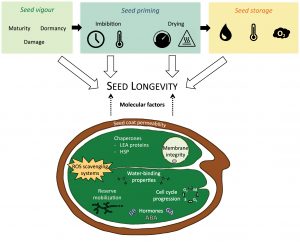 Seed priming is a family of presowing techniques that control water uptake in seeds, so they are dehydrated once metabolic activities have restarted but before the end of germination. Primed seeds have shown increased and more synchronized germination, especially under stressful conditions, making these techniques highly interesting to be applied in agriculture. However, its impact on seed longevity –a fundamental trait for seed management and conservation– is highly variable. Here, Fabrissin and colleagues synthesize the effect of different priming parameters on seed longevity to provide guidelines to properly exploit this technique’s benefits. Overall, primed seeds have reduced seed longevity when compared to non-treated seeds. However, different results were found when seeds of the same species were treated with different techniques such a hydropriming (soaking under optimal temperatures), osmopriming (exposure to desiccant solutions), or hormopriming (imbibition on plant growth regulators). Different molecular pathways were induced by different priming treatments, suggesting the activation of distinct mechanisms that affect seed longevity. Current evidence suggests that adverse effects on seed longevity could be minimized by manipulating various priming parameters, such as imbibition, temperature, and posterior storage conditions. As a result, this review provides an exciting starting point for future research that could lead to widespread use of seed priming without compromising seed lifespan. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Exp. Bot. 10.1093/jxb/erab004
Seed priming is a family of presowing techniques that control water uptake in seeds, so they are dehydrated once metabolic activities have restarted but before the end of germination. Primed seeds have shown increased and more synchronized germination, especially under stressful conditions, making these techniques highly interesting to be applied in agriculture. However, its impact on seed longevity –a fundamental trait for seed management and conservation– is highly variable. Here, Fabrissin and colleagues synthesize the effect of different priming parameters on seed longevity to provide guidelines to properly exploit this technique’s benefits. Overall, primed seeds have reduced seed longevity when compared to non-treated seeds. However, different results were found when seeds of the same species were treated with different techniques such a hydropriming (soaking under optimal temperatures), osmopriming (exposure to desiccant solutions), or hormopriming (imbibition on plant growth regulators). Different molecular pathways were induced by different priming treatments, suggesting the activation of distinct mechanisms that affect seed longevity. Current evidence suggests that adverse effects on seed longevity could be minimized by manipulating various priming parameters, such as imbibition, temperature, and posterior storage conditions. As a result, this review provides an exciting starting point for future research that could lead to widespread use of seed priming without compromising seed lifespan. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) J. Exp. Bot. 10.1093/jxb/erab004
Viewpoint: Bryophytes are not early diverging land plants
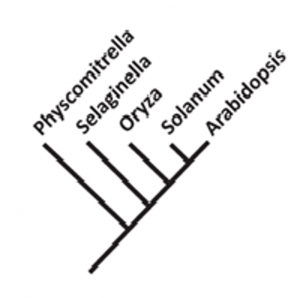
Phylogenetic trees demonstrate the evolutionary relationship between organisms, but sometimes these relationships are described by inappropriate terminology. In this Viewpoint, McDaniel observes that based on a phylogeny as drawn here, bryophytes (represented by the genus Physcomitrella, aka Physcomitrium) are frequently described as an early-diverging, ancient, or basal lineage in relation to the rest of terrestrial plants. But this terminology suggests that evolution has a direction towards the state of a higher lineage, and also suggests that bryophytes stopped evolving after diverging and maintained their ancestral traits. Furthermore, it implies a sort of hierarchy as in Aristotle’s Scala Naturae – in which things are ranked from low (rocks) to high (angels), which McDaniel describes as “pre-evolutionary” thinking. The author argues that we should instead use more accurate language such as “sister group” or “common ancestor” when describing relationships. Finally, the author emphasizes how using proper language improves the perception of students, the public, and the scientists outside the field on modern evolutionary concepts (he provides links to some excellent resources for teaching this “tree-thinking” perspective). Proper language could better orient research questions and prevent research bias towards so-called “advanced lineages.” All living organisms are still evolving; bryophytes and angiosperms alike. (Summary by Roxana Khoshravesh @RoxiKh) New Phytol. 10.1111/nph.17241
An evidence-based 3D reconstruction of Asteroxylon mackiei, the most complex plant preserved from the Rhynie chert
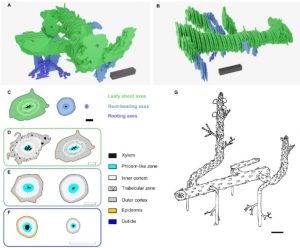 The Rhynie chert holds some of the oldest and best-preserved fossils of vascular plants (Rhynie is a village in north-east Scotland, and chert is a type of sedimentary rock). Analysis of thin section of fossils from here have provided the earliest cellular details of “rhizoid-based rooting systems, germinating spores and fossilised meristems.” But as Hetherington et al. observe, it is difficult to reconstruct overall body plans from thin sections. Here they have examined serial thick as well as thin sections of two complete lycopsid Asteroxylon mackiei plants, which are among the earliest group of land plants in the fossil record with complex body plans. Their findings demonstrate that the rooting system of these plants branched differently than what is observed in extant lycopsids. Specifically, in today’s plants the roots originate endogenously – the meristem of the new axis develops within the tissue of the parent axis and breaks through it. By contrast, the fossil plants show evidence of dichotomous branching (in which the tip of the parent axes splits), specifically anisotomous dichotomy meaning that the split is unequal. This type of root development, not found in extant lycopsids, is now extinct. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2021.04.11.439326
The Rhynie chert holds some of the oldest and best-preserved fossils of vascular plants (Rhynie is a village in north-east Scotland, and chert is a type of sedimentary rock). Analysis of thin section of fossils from here have provided the earliest cellular details of “rhizoid-based rooting systems, germinating spores and fossilised meristems.” But as Hetherington et al. observe, it is difficult to reconstruct overall body plans from thin sections. Here they have examined serial thick as well as thin sections of two complete lycopsid Asteroxylon mackiei plants, which are among the earliest group of land plants in the fossil record with complex body plans. Their findings demonstrate that the rooting system of these plants branched differently than what is observed in extant lycopsids. Specifically, in today’s plants the roots originate endogenously – the meristem of the new axis develops within the tissue of the parent axis and breaks through it. By contrast, the fossil plants show evidence of dichotomous branching (in which the tip of the parent axes splits), specifically anisotomous dichotomy meaning that the split is unequal. This type of root development, not found in extant lycopsids, is now extinct. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2021.04.11.439326
Trapped in traffic – redefining cellular responses to auxin pharmacology
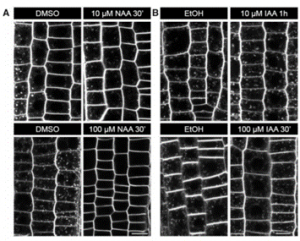 Understanding cellular responses to the phytohormone auxin has always been a challenge and decades of work has constantly renewed our knowledge of the same. Here, Narasimhan and colleagues attempted to revisit and resolve the basis of differential action of naturally occurring auxin indole-3-acetic acid (IAA) and its synthetic analogue naphthalene acetic acid (NAA) in mediating intracellular trafficking processes. Replicating classical experiments to understand the mechanism of auxin inhibition of the action of fungal toxin Brefeldin-A (BFA), they found both IAA and NAA to non-specifically inhibit formation of the endosomal aggregations. However, only NAA interfered in the endocytic recycling of proteins by modifying the lipid profile, and by effect the structure, of the endomembrane system. Interestingly, NAA and IAA acted alike to promote the specific, clathrin-mediated endocytosis of auxin efflux carrier PIN2, which the authors found to be necessary for its polar localization. Together, this study highlights the need to employ non-BFA based experiments to infer auxin effect on endocytosis and builds the foundation to better understand cellular events influenced by the pharmacological application of auxin analogues. (Summary by Pavithran Narayanan @pavi_narayanan) Plant Physiol. 10.1093/plphys/kiab134. Note: The implications of these findings are far-ranging and discussed in three accompanying commentaries that can be found here, here and here.
Understanding cellular responses to the phytohormone auxin has always been a challenge and decades of work has constantly renewed our knowledge of the same. Here, Narasimhan and colleagues attempted to revisit and resolve the basis of differential action of naturally occurring auxin indole-3-acetic acid (IAA) and its synthetic analogue naphthalene acetic acid (NAA) in mediating intracellular trafficking processes. Replicating classical experiments to understand the mechanism of auxin inhibition of the action of fungal toxin Brefeldin-A (BFA), they found both IAA and NAA to non-specifically inhibit formation of the endosomal aggregations. However, only NAA interfered in the endocytic recycling of proteins by modifying the lipid profile, and by effect the structure, of the endomembrane system. Interestingly, NAA and IAA acted alike to promote the specific, clathrin-mediated endocytosis of auxin efflux carrier PIN2, which the authors found to be necessary for its polar localization. Together, this study highlights the need to employ non-BFA based experiments to infer auxin effect on endocytosis and builds the foundation to better understand cellular events influenced by the pharmacological application of auxin analogues. (Summary by Pavithran Narayanan @pavi_narayanan) Plant Physiol. 10.1093/plphys/kiab134. Note: The implications of these findings are far-ranging and discussed in three accompanying commentaries that can be found here, here and here.
Structural insights into photosystem II assembly
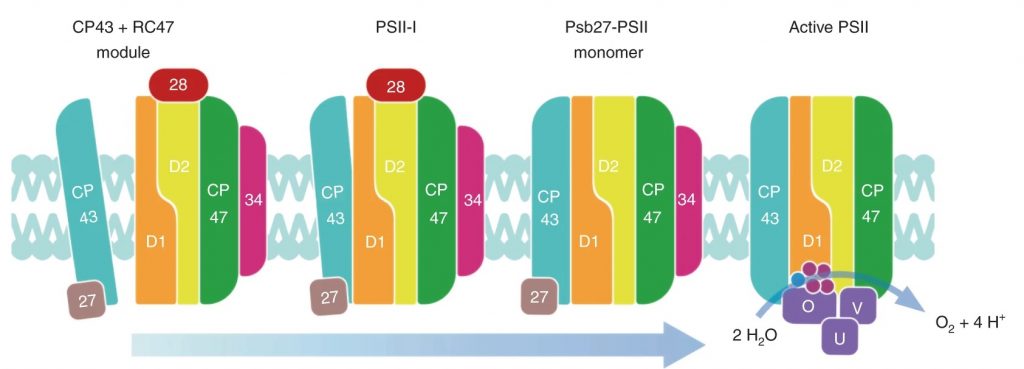 Photosystem II is a multisubunit enzyme complex in the thylakoid membranes of plant, algae, and cyanobacteria. Here, Zebret et al. used cryo-electron microscopy to produce high-resolution structural information of the intermediate complexes involved in the biogenesis and assembly of photosystem II from the cyanobacterium Thermosynechococcus elongatus. This study shows how the spatiotemporally synchronized biogenesis process is facilitated by a network of assembly factors, particularly Psb27, Psb28 and Psb34, and reveals an important transition phase before the activation of the oxygen evolving complex (OEC). At the future site where the OEC will bind, there is a single positively charged ion that coordinates with the three amino acids Asp170, Glu189 and His332 of D1. As the authors suggest, this site “seems to be prepared to accept the Mn4CaO5 cluster, as indicated by the similarities in side-chain positioning.” (Summary by Arun K. Shanker @arunshanker) Nature Plants 10.1038/s41477-021-00895-0
Photosystem II is a multisubunit enzyme complex in the thylakoid membranes of plant, algae, and cyanobacteria. Here, Zebret et al. used cryo-electron microscopy to produce high-resolution structural information of the intermediate complexes involved in the biogenesis and assembly of photosystem II from the cyanobacterium Thermosynechococcus elongatus. This study shows how the spatiotemporally synchronized biogenesis process is facilitated by a network of assembly factors, particularly Psb27, Psb28 and Psb34, and reveals an important transition phase before the activation of the oxygen evolving complex (OEC). At the future site where the OEC will bind, there is a single positively charged ion that coordinates with the three amino acids Asp170, Glu189 and His332 of D1. As the authors suggest, this site “seems to be prepared to accept the Mn4CaO5 cluster, as indicated by the similarities in side-chain positioning.” (Summary by Arun K. Shanker @arunshanker) Nature Plants 10.1038/s41477-021-00895-0
DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis
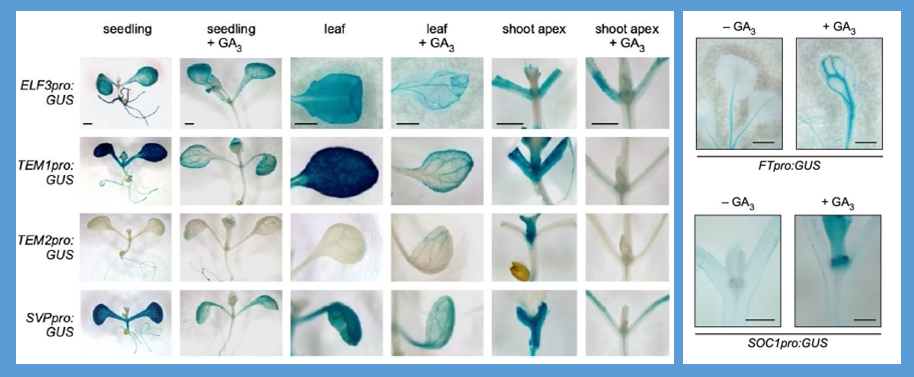 In Arabidopsis, flowering is promoted by the activation of the florigen FT in response to long days and by the accumulation of the phytohormone GA under short days (SD). Despite the extensive knowledge of regulatory networks involving FT, the molecular mechanism controlled by GA is still unresolved. Fukazawa and coworkers previously identified GAI-ASSOCIATED FACTOR1 (GAF1), a transcription factor that interacts with the growth repressors DELLA and the corepressor TOPLESS RELATED (TPR), and found that GAs convert the GAF1 complex from a transcriptional activator to a repressor via the degradation of DELLAs. In addition, they proposed a specific role for GAF1 in the regulation of flowering mediated by GA, supported by the late flowering phenotype of gaf1 mutants under SD. In this study, the authors used RNA-sequencing to identify GAF1 targets and found mis-regulation of ELF3, SVP and TEM1/TEM2 genes, which encode important floral repressors. Specifically, these flowering time genes were downregulated upon GAF1 induction and their regulatory sequences were directly bound by GAF1. Furthermore, application of GAs caused significant reduction of ELF3, SVP and TEM1/TEM2 expression in wildtypes but not in gaf1 mutants. Taken together, these findings suggested that GAs cause downregulation of GAF1 target genes, which leads to the induction of FT in the leaf and SOC1 in the shoot apical meristem. To recap, GAF1 forms a repressor complex with TRP in response to accumulation of GAs, which occurs shortly before the floral transition under SD, that switches off key floral repressors and switches on floral integrators. (Summary and image adaptation by Michela Osnato @michela_osnato) Plant Cell 10.1093/plcell/koab102
In Arabidopsis, flowering is promoted by the activation of the florigen FT in response to long days and by the accumulation of the phytohormone GA under short days (SD). Despite the extensive knowledge of regulatory networks involving FT, the molecular mechanism controlled by GA is still unresolved. Fukazawa and coworkers previously identified GAI-ASSOCIATED FACTOR1 (GAF1), a transcription factor that interacts with the growth repressors DELLA and the corepressor TOPLESS RELATED (TPR), and found that GAs convert the GAF1 complex from a transcriptional activator to a repressor via the degradation of DELLAs. In addition, they proposed a specific role for GAF1 in the regulation of flowering mediated by GA, supported by the late flowering phenotype of gaf1 mutants under SD. In this study, the authors used RNA-sequencing to identify GAF1 targets and found mis-regulation of ELF3, SVP and TEM1/TEM2 genes, which encode important floral repressors. Specifically, these flowering time genes were downregulated upon GAF1 induction and their regulatory sequences were directly bound by GAF1. Furthermore, application of GAs caused significant reduction of ELF3, SVP and TEM1/TEM2 expression in wildtypes but not in gaf1 mutants. Taken together, these findings suggested that GAs cause downregulation of GAF1 target genes, which leads to the induction of FT in the leaf and SOC1 in the shoot apical meristem. To recap, GAF1 forms a repressor complex with TRP in response to accumulation of GAs, which occurs shortly before the floral transition under SD, that switches off key floral repressors and switches on floral integrators. (Summary and image adaptation by Michela Osnato @michela_osnato) Plant Cell 10.1093/plcell/koab102
Pollen PCP-B peptides unlock a stigma peptide–receptor kinase gating mechanism for pollination
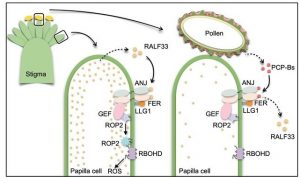 It’s spring where I live and the air and my eyes are filled with pollen. The lucky few pollen grains that land on the stigmatic surface of a flower might germinate and deliver the sperm to the ovule, but there are many checkpoints even after their safe landing. Here, Liu et al. uncover one of the earliest tests that must be passed for pollen to germinate. Previous studies have shown an important role for small peptides as signals during pollination. In this new work, the authors found that one of these is necessary for pollen to hydrate on the stigmatic surface. They showed that the peptide produced by the pollen competes with a stigma-produced peptide for biding to a receptor-like kinase. When the pollen peptide binds, levels of reactive oxygen species in the stigma drop, allowing the pollen to hydrate, moving one step closer to successful fertilization. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.abc6107
It’s spring where I live and the air and my eyes are filled with pollen. The lucky few pollen grains that land on the stigmatic surface of a flower might germinate and deliver the sperm to the ovule, but there are many checkpoints even after their safe landing. Here, Liu et al. uncover one of the earliest tests that must be passed for pollen to germinate. Previous studies have shown an important role for small peptides as signals during pollination. In this new work, the authors found that one of these is necessary for pollen to hydrate on the stigmatic surface. They showed that the peptide produced by the pollen competes with a stigma-produced peptide for biding to a receptor-like kinase. When the pollen peptide binds, levels of reactive oxygen species in the stigma drop, allowing the pollen to hydrate, moving one step closer to successful fertilization. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.abc6107
SeedQuant: A deep learning-based tool for assessing stimulant and inhibitor activity on root parasitic seeds
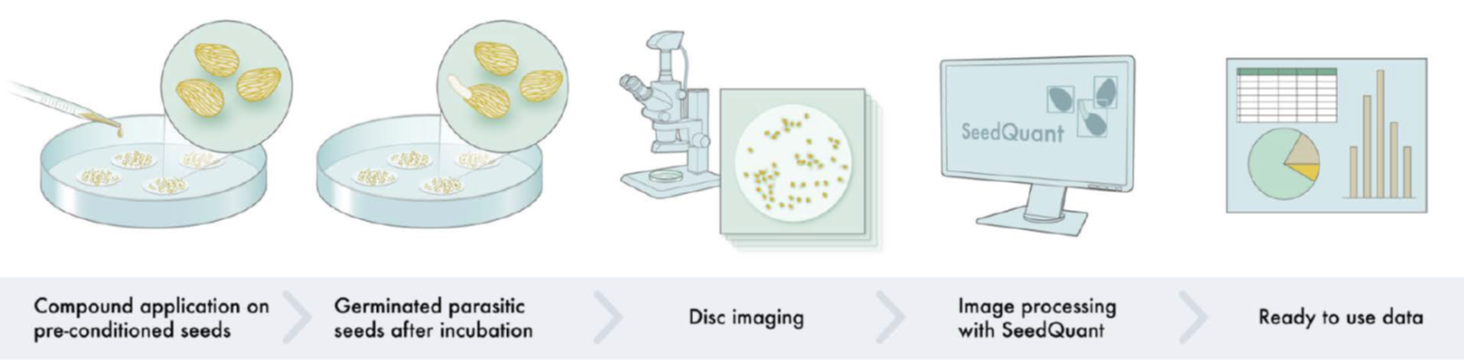 Parasitic plants can completely wipe out a farmer’s harvest. The seeds can lie dormant in soil until they perceive signals from potential host plants, which stimulate germination. Therefore, germination stimulants applied to fields before the crops are planted can cause the parasitic seeds to germinate and die in the absence of their host. Many compounds have been studied to try to identify cheap, effective, stable germination stimulants, but the scoring of their effect (counting seed germination) is time consuming. Here, Braguy, Ramazanova, Giancola et al. describe a tool, created by deep learning (a subfield of artificial intelligence) that is able to quickly and efficiently quantify seed germination. The tool, which uses a Faster Region-based Convolutional Neural Network (Faster R-CNN) to assess seeds, is 95% accurate and requires only a fraction of a second to extract data. SeedQuant is free to use and can be found here https://braguyjm.github.io/SeedQuant2. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab173
Parasitic plants can completely wipe out a farmer’s harvest. The seeds can lie dormant in soil until they perceive signals from potential host plants, which stimulate germination. Therefore, germination stimulants applied to fields before the crops are planted can cause the parasitic seeds to germinate and die in the absence of their host. Many compounds have been studied to try to identify cheap, effective, stable germination stimulants, but the scoring of their effect (counting seed germination) is time consuming. Here, Braguy, Ramazanova, Giancola et al. describe a tool, created by deep learning (a subfield of artificial intelligence) that is able to quickly and efficiently quantify seed germination. The tool, which uses a Faster Region-based Convolutional Neural Network (Faster R-CNN) to assess seeds, is 95% accurate and requires only a fraction of a second to extract data. SeedQuant is free to use and can be found here https://braguyjm.github.io/SeedQuant2. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab173



