Plant Science Reseach Weekly: March 22nd
Review: Molecular networks of seed size control in plants ($)
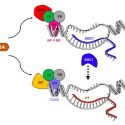
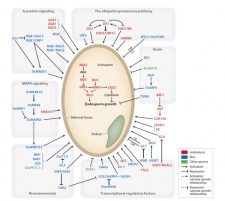 Crop yield is largely determined by the size of seeds, and studies are being conducted to understand the complex molecular network controlling the seed size. Li et al. review the possible molecular mechanisms and regulatory networks underlying seed size control and growth, including the factors originating in both maternal and zygotic tissues. The authors describe several pathways involved in the control of seed size including: the ubiquitin-proteasome pathway, G-proteins, MAP kinases, and the HAIKU pathway, which together with phytohomornes influences embryo development. Studies on the role of the embryo in determining seed size and factors controlling embryo development are limited. Currently, delineating the upstream and downstream components of known seed size regulators and their interplay is necessary to understand the complete circuitry governing seed/grain size. Given that the mechanisms of seed size control are largely conserved, the research should now focus on translating the knowledge generated in model plants to improve seed size in cultivated crops. (Summary by Muthamilarasan Mehanathan) Annu Rev Plant Biol 10.1146/annurev-arplant-050718-095851
Crop yield is largely determined by the size of seeds, and studies are being conducted to understand the complex molecular network controlling the seed size. Li et al. review the possible molecular mechanisms and regulatory networks underlying seed size control and growth, including the factors originating in both maternal and zygotic tissues. The authors describe several pathways involved in the control of seed size including: the ubiquitin-proteasome pathway, G-proteins, MAP kinases, and the HAIKU pathway, which together with phytohomornes influences embryo development. Studies on the role of the embryo in determining seed size and factors controlling embryo development are limited. Currently, delineating the upstream and downstream components of known seed size regulators and their interplay is necessary to understand the complete circuitry governing seed/grain size. Given that the mechanisms of seed size control are largely conserved, the research should now focus on translating the knowledge generated in model plants to improve seed size in cultivated crops. (Summary by Muthamilarasan Mehanathan) Annu Rev Plant Biol 10.1146/annurev-arplant-050718-095851
Q&A: Modern crop breeding for future food security
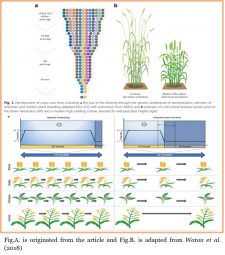 The demand for plant-based products will increase and must be doubled in the near future. Therefore, there is need for technological advancements and skillsets in all fields related to agriculture to successfully produce more efficiently than now. What could be done to speed up production to meet the future demand of food? Voss-Fell et al. address this issue in a Q&A (Questions and answers) and discuss how new technologies in plant breeding could be a fast-track for the development of new varieties, and therefore, to meet future plant-based product demands. Genomic selection, gene editing (using CRISPR-Cas genetic engineering systems), mutagenesis-based breeding and the current method as alternative for gene editing or gene introgression called speed breeding are some of the technologies developed as crop breeding directed-methods. The speed breeding approach developed by Dr. Lee Hickey and colleagues in the fast-forwarding genetic gain can speed and reduce the number of generations up to six per year for some crops. Thus, an integrated approach of more efficient breeding strategies using a combination of these new technologies could pave the way to plant-based food development and thus meet the food demands. (Summary by Florian Ahloumessou) BMC Biology. 10.1186/s12915-019-0638-4
The demand for plant-based products will increase and must be doubled in the near future. Therefore, there is need for technological advancements and skillsets in all fields related to agriculture to successfully produce more efficiently than now. What could be done to speed up production to meet the future demand of food? Voss-Fell et al. address this issue in a Q&A (Questions and answers) and discuss how new technologies in plant breeding could be a fast-track for the development of new varieties, and therefore, to meet future plant-based product demands. Genomic selection, gene editing (using CRISPR-Cas genetic engineering systems), mutagenesis-based breeding and the current method as alternative for gene editing or gene introgression called speed breeding are some of the technologies developed as crop breeding directed-methods. The speed breeding approach developed by Dr. Lee Hickey and colleagues in the fast-forwarding genetic gain can speed and reduce the number of generations up to six per year for some crops. Thus, an integrated approach of more efficient breeding strategies using a combination of these new technologies could pave the way to plant-based food development and thus meet the food demands. (Summary by Florian Ahloumessou) BMC Biology. 10.1186/s12915-019-0638-4
High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants
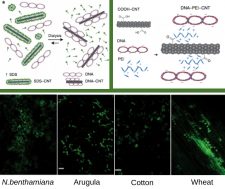 Key to success of crop improvement is the development of easier, faster and safer biomolecules-delivery systems. Here, the main limitation is the cell wall, which compromises the yield of exogenic material transfer to plant cells. In this study, Demirer et al demonstrate the advantages of infiltrated DNA-nanocarriers in monocot and dicot plants leaves (Nicotiana benthamiana, arugula, wheat and cotton) and arugula protoplasts. The authors developed two grafting methods for loading reporter GFP-encoding plasmids and linear amplicons on single and multi-walled carbon nanotubes (CNTs): using dialysis by coating CNTs with a surfactant (SDS), and using electrostatic interactions onto positively-charged, poly-ethylenimine (PEI)-coated CNTs. Internalization in the plant cell cytosol and nucleus was confirmed using Cy3-tagged DNA-CNTs with confocal microscopy, transmission electron microscopy, and near-infrared (NIR) imaging (CNTs have intrinsic NIR fluorescence). Quantitative PCR and droplet digital PCR (ddPCR) analysis showed that GFP expression is transient, which satisfies GMO regulations. Additionally, qPCR analysis of a stress gene and photosystem II quantum yield measurements revealed no sign of induced toxicity by the nanocarriers. Thus, the authors suggest CNT-mediated delivery is a highly efficient and species-independent tool, which “enable high-throughput plant genetic transformations”. (Summary by Ana Valladares) Nature Nanotechnology 10.1038/s41565-019-0382-5
Key to success of crop improvement is the development of easier, faster and safer biomolecules-delivery systems. Here, the main limitation is the cell wall, which compromises the yield of exogenic material transfer to plant cells. In this study, Demirer et al demonstrate the advantages of infiltrated DNA-nanocarriers in monocot and dicot plants leaves (Nicotiana benthamiana, arugula, wheat and cotton) and arugula protoplasts. The authors developed two grafting methods for loading reporter GFP-encoding plasmids and linear amplicons on single and multi-walled carbon nanotubes (CNTs): using dialysis by coating CNTs with a surfactant (SDS), and using electrostatic interactions onto positively-charged, poly-ethylenimine (PEI)-coated CNTs. Internalization in the plant cell cytosol and nucleus was confirmed using Cy3-tagged DNA-CNTs with confocal microscopy, transmission electron microscopy, and near-infrared (NIR) imaging (CNTs have intrinsic NIR fluorescence). Quantitative PCR and droplet digital PCR (ddPCR) analysis showed that GFP expression is transient, which satisfies GMO regulations. Additionally, qPCR analysis of a stress gene and photosystem II quantum yield measurements revealed no sign of induced toxicity by the nanocarriers. Thus, the authors suggest CNT-mediated delivery is a highly efficient and species-independent tool, which “enable high-throughput plant genetic transformations”. (Summary by Ana Valladares) Nature Nanotechnology 10.1038/s41565-019-0382-5
Life between the cells: measuring apoplast hydration and content
 The extracellular space within the leaves, or apoplast, has an active role in many aspects of plant physiology; since water from transpiration, sucrose from photosynthesis and other metabolites pass through the apoplast. Since abiotic and biotic factors (i.e., pathogens) modify the composition of the apoplast, several methods have been developed to study its content. In a recent report, Gentzel et al. refine a method based in infiltration-centrifugation that allows apoplast fluid extraction and rapid estimation of apoplast hydration. To measure cell damage during the procedure, they tested wash solutions, centrifugation force and evaluated cell damage using confocal microscopy. As an application of the method, the authors quantified the water soaking response to pathogens in maize plants. Further applications of this method could include expression analysis in plant-pathogen interactions and analysis of metabolism. (Summary by Humberto Herrera-Ubaldo). Plant Physiol. 10.1104/pp.18.01076
The extracellular space within the leaves, or apoplast, has an active role in many aspects of plant physiology; since water from transpiration, sucrose from photosynthesis and other metabolites pass through the apoplast. Since abiotic and biotic factors (i.e., pathogens) modify the composition of the apoplast, several methods have been developed to study its content. In a recent report, Gentzel et al. refine a method based in infiltration-centrifugation that allows apoplast fluid extraction and rapid estimation of apoplast hydration. To measure cell damage during the procedure, they tested wash solutions, centrifugation force and evaluated cell damage using confocal microscopy. As an application of the method, the authors quantified the water soaking response to pathogens in maize plants. Further applications of this method could include expression analysis in plant-pathogen interactions and analysis of metabolism. (Summary by Humberto Herrera-Ubaldo). Plant Physiol. 10.1104/pp.18.01076
Rapid evolution of protein diversity by de novo origination in Oryza
 The de-novo origin of new protein-coding genes from non-coding regions of plant genomes is a contributor to protein diversity, although it has been difficult to quantify to what extent this process occurs. High quality reference genome maps, deep transcriptome and targeted proteome sequencing are requisite for discovery of de novo genes. Using genome and transcriptome data from 10 members of genus Oryza, Zhang et al. identified 175 de novo originating open reading frames. The origin of new genes was mapped and the molecular events identified that led to their emergence, such as indels and premature termination codon disappearance. Interestingly, several of these new genes that were validated to form an active protein express in a more tissue specific manner than comparable old singleton genes. (Summary by Kaushal Kumar Bhati) Nature Ecol. Evol. 10.1038/s41559-019-0822-5
The de-novo origin of new protein-coding genes from non-coding regions of plant genomes is a contributor to protein diversity, although it has been difficult to quantify to what extent this process occurs. High quality reference genome maps, deep transcriptome and targeted proteome sequencing are requisite for discovery of de novo genes. Using genome and transcriptome data from 10 members of genus Oryza, Zhang et al. identified 175 de novo originating open reading frames. The origin of new genes was mapped and the molecular events identified that led to their emergence, such as indels and premature termination codon disappearance. Interestingly, several of these new genes that were validated to form an active protein express in a more tissue specific manner than comparable old singleton genes. (Summary by Kaushal Kumar Bhati) Nature Ecol. Evol. 10.1038/s41559-019-0822-5
A large‐scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis ($)
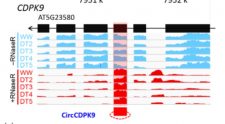 Recently, a novel class of noncoding RNAs named circular RNA (circRNA) has been shown to play roles in transcriptional regulation of gene expression in plants. However, a complete picture of circRNAome and their regulatory roles in controlling the stress response of plants was lacking. To address this, Zhang et al. have performed circRNA-seq and mRNA-seq of wild-type maize (B73) and Arabidopsis (Col) subjected to well-watered and drought-treated conditions. The analysis revealed the presence of 2174 and 1354 high-confidence circRNAs in maize and Arabidopsis, respectively, and the expression of circRNA-hosting genes was found to be significantly higher than the other coding genes. Correlating the expression of circular and linear transcripts in drought response showed similar variation patterns in maize and Arabidopsis suggesting the conserved molecular mechanisms governing drought response in plants. Investigating the effect of circRNAs on smallRNA (sRNA) in maize implicated a negative correlation between circRNA expression and sRNA accumulation. Further, overexpression of a circular RNA (circGORK) hosted by the GORK (Guard cell outward-rectifying K+-channel) gene in Arabidopsis indicated a positive role of circGORK in conferring drought tolerance. The data suggest that circGORK may influence the expression/splicing of its cognate linear transcripts. Altogether, the study delineates the involvement of circRNA in regulating drought stress response, and also, provides the framework for further research in the area of circRNA-mediated stress response. (Summary by Muthamilarasan Mehanathan) Plant J 10.1111/tpj.14267
Recently, a novel class of noncoding RNAs named circular RNA (circRNA) has been shown to play roles in transcriptional regulation of gene expression in plants. However, a complete picture of circRNAome and their regulatory roles in controlling the stress response of plants was lacking. To address this, Zhang et al. have performed circRNA-seq and mRNA-seq of wild-type maize (B73) and Arabidopsis (Col) subjected to well-watered and drought-treated conditions. The analysis revealed the presence of 2174 and 1354 high-confidence circRNAs in maize and Arabidopsis, respectively, and the expression of circRNA-hosting genes was found to be significantly higher than the other coding genes. Correlating the expression of circular and linear transcripts in drought response showed similar variation patterns in maize and Arabidopsis suggesting the conserved molecular mechanisms governing drought response in plants. Investigating the effect of circRNAs on smallRNA (sRNA) in maize implicated a negative correlation between circRNA expression and sRNA accumulation. Further, overexpression of a circular RNA (circGORK) hosted by the GORK (Guard cell outward-rectifying K+-channel) gene in Arabidopsis indicated a positive role of circGORK in conferring drought tolerance. The data suggest that circGORK may influence the expression/splicing of its cognate linear transcripts. Altogether, the study delineates the involvement of circRNA in regulating drought stress response, and also, provides the framework for further research in the area of circRNA-mediated stress response. (Summary by Muthamilarasan Mehanathan) Plant J 10.1111/tpj.14267
The RNA export factor ALY1 enables genome-wide RNA-directed DNA methylation
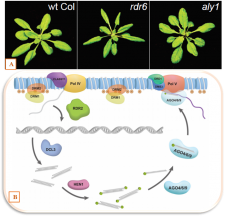 The epigenetic control of gene expression is crucial for genomic stability and allows the defence against invading DNA that for instance could be derived from viruses or transposable elements. In this context, the RNA-directed DNA methylation (RdDM) plays an essential role in silencing of genes via epigenetically modifying the DNA. Here, small RNA molecules direct a protein complex to a target region that finally catalyses the methylation of cysteine residues thus leading to a transcriptional repression. The RdDM-pathways involves several components including the endoribonuclease DICER-like 3, the ARGONAUTE proteins 4 and 6 as well as the RNA Polymerase V and methylases. Choudury et al. describe an Arabidopsis thaliana mutant that is deficient in the RdDM-mechanism and thus vulnerable to the expression of foreign DNA. They identify the corresponding gene, ALY1, which is involved in the export of RNA from the nucleus. In aly1, the mRNA of several factors such as ARGONAUTE6 and Pol V cannot be efficiently exported leading to a decreased abundance of these proteins thus to a damaged RdDM-pathway. (Summary by Florian Ahloumessou) Plant Cell 10.1105/tpc.18.00624
The epigenetic control of gene expression is crucial for genomic stability and allows the defence against invading DNA that for instance could be derived from viruses or transposable elements. In this context, the RNA-directed DNA methylation (RdDM) plays an essential role in silencing of genes via epigenetically modifying the DNA. Here, small RNA molecules direct a protein complex to a target region that finally catalyses the methylation of cysteine residues thus leading to a transcriptional repression. The RdDM-pathways involves several components including the endoribonuclease DICER-like 3, the ARGONAUTE proteins 4 and 6 as well as the RNA Polymerase V and methylases. Choudury et al. describe an Arabidopsis thaliana mutant that is deficient in the RdDM-mechanism and thus vulnerable to the expression of foreign DNA. They identify the corresponding gene, ALY1, which is involved in the export of RNA from the nucleus. In aly1, the mRNA of several factors such as ARGONAUTE6 and Pol V cannot be efficiently exported leading to a decreased abundance of these proteins thus to a damaged RdDM-pathway. (Summary by Florian Ahloumessou) Plant Cell 10.1105/tpc.18.00624
Four auxin-like compounds can selectively regulate plant development
 Plants have developed their own strategies to ensure developmental plasticity, such as the use of phytohormones. The phytohormone auxin affects multiple aspects of plant development. Auxin is perceived via a co-receptor complex called TIR1/AFB-AUX/IAA, and SCF TIR1/AFB degrades AUX/IAA proteins to liberate the downstream transcription factors, leading to the auxin response. However, the degradation rate of various AUX/IAAs differs a lot. To better characterize the complicated auxin response, Vain et al. have isolated and characterized four auxin agonists (RN1 to 4) from a chemical biology screen. These molecules can selectively promote or inhibit AUX/IAA degradation in a dose-dependent manner, contributing to specific auxin responses at transcriptional, biochemical and morphological levels. The discovery of auxinic compounds is helpful for us to study the specific mechanisms of auxin response in Arabidopsis, and also beneficial to develop agrochemicals to boost agriculture and forestry for the world. (Summary by Nanxun Qin) PNAS 10.1073/pnas.1809037116
Plants have developed their own strategies to ensure developmental plasticity, such as the use of phytohormones. The phytohormone auxin affects multiple aspects of plant development. Auxin is perceived via a co-receptor complex called TIR1/AFB-AUX/IAA, and SCF TIR1/AFB degrades AUX/IAA proteins to liberate the downstream transcription factors, leading to the auxin response. However, the degradation rate of various AUX/IAAs differs a lot. To better characterize the complicated auxin response, Vain et al. have isolated and characterized four auxin agonists (RN1 to 4) from a chemical biology screen. These molecules can selectively promote or inhibit AUX/IAA degradation in a dose-dependent manner, contributing to specific auxin responses at transcriptional, biochemical and morphological levels. The discovery of auxinic compounds is helpful for us to study the specific mechanisms of auxin response in Arabidopsis, and also beneficial to develop agrochemicals to boost agriculture and forestry for the world. (Summary by Nanxun Qin) PNAS 10.1073/pnas.1809037116
Regulation of seed dormancy by ETR1/RDO3
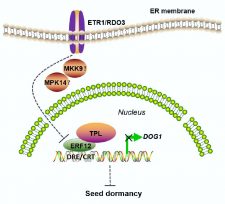 Seed dormancy is an essential fitness trait for plants as it allows their seeds to survive adverse seasons and to synchronise their germination with the occurrence of suitable conditions. While the molecular pathways of the major phytohormones involved in seed dormancy have been largely elucidated, the mechanisms of others remain elusive. In the case of ethylene the underlying molecular mechanism is not well understood although this phytohormone has been shown to play a pivotal role in the control of seed dormancy and germination. Here, Li et al. report the identification of the Arabidopsis reduced dormancy 3 (rdo3) mutant and show that RDO3 encodes the ethylene receptor ETR1. Furthermore, they demonstrate that ERF12 plays a key role in the regulation of seed dormancy by interacting with the transcriptional co-repressor TOPLESS (TPL) in the downstream pathway of ethylene signaling mediated by ETR1/RDO3. Moreover, major dormancy factor DELAY OF GERMINATION1 (DOG1) is a direct target of the ERF12-85-TPL repressor complex, underlining the role of ethylene signaling in the regulation of seed dormancy. (Summary by Florian Ahloumessou) Plant Cell 10.1105/tpc.18.00449
Seed dormancy is an essential fitness trait for plants as it allows their seeds to survive adverse seasons and to synchronise their germination with the occurrence of suitable conditions. While the molecular pathways of the major phytohormones involved in seed dormancy have been largely elucidated, the mechanisms of others remain elusive. In the case of ethylene the underlying molecular mechanism is not well understood although this phytohormone has been shown to play a pivotal role in the control of seed dormancy and germination. Here, Li et al. report the identification of the Arabidopsis reduced dormancy 3 (rdo3) mutant and show that RDO3 encodes the ethylene receptor ETR1. Furthermore, they demonstrate that ERF12 plays a key role in the regulation of seed dormancy by interacting with the transcriptional co-repressor TOPLESS (TPL) in the downstream pathway of ethylene signaling mediated by ETR1/RDO3. Moreover, major dormancy factor DELAY OF GERMINATION1 (DOG1) is a direct target of the ERF12-85-TPL repressor complex, underlining the role of ethylene signaling in the regulation of seed dormancy. (Summary by Florian Ahloumessou) Plant Cell 10.1105/tpc.18.00449
Constitutive signaling activity of a receptor-associated protein links fertilization with embryonic patterning in Arabidopsis thaliana ($)
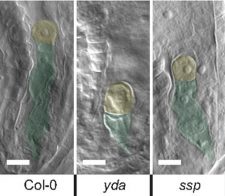 The apical and basal identity of a growing embryo is determined by an asymmetrical division of the zygote in flowering plants. The MAPKK kinase YODA (YDA) is important for zygotic elongation and embryonic polarity. During embryo development, YDA is activated by the membrane associated pseudo-kinase SHORT SUSPENSOR (SSP), which belongs to BRASSINOSTEROID SIGNALING KINASE (BSK) family and expresses specifically in sperm cells of pollen. SSP is unusual in that it constitutively activates YDA. The authors’ studies suggest that the delivery of SSP during fertilization is sufficient to activate YDA and establish apical-basal polarity in the embryo. By losing a regulatory interaction SSP can constitutively activates YDA. Additionally, BSK1 and BSK2 work independently to SSP to genetically control the YDA role in early embryonic development. (Summary by Kaushal Kumar Bhati) PNAS
The apical and basal identity of a growing embryo is determined by an asymmetrical division of the zygote in flowering plants. The MAPKK kinase YODA (YDA) is important for zygotic elongation and embryonic polarity. During embryo development, YDA is activated by the membrane associated pseudo-kinase SHORT SUSPENSOR (SSP), which belongs to BRASSINOSTEROID SIGNALING KINASE (BSK) family and expresses specifically in sperm cells of pollen. SSP is unusual in that it constitutively activates YDA. The authors’ studies suggest that the delivery of SSP during fertilization is sufficient to activate YDA and establish apical-basal polarity in the embryo. By losing a regulatory interaction SSP can constitutively activates YDA. Additionally, BSK1 and BSK2 work independently to SSP to genetically control the YDA role in early embryonic development. (Summary by Kaushal Kumar Bhati) PNAS
Root branching is not induced by auxins in Selaginella moellendorffii
 Auxin is crucial for root development across angiosperms. All the auxin signal transduction pathway components are conserved in land plants, but their role is still not completely clear in early diverged lineages. Here, Fang et al. present a comprehensive analysis of exogenous auxin addition and polar transport inhibitor effects in the root development of Selaginella moellendorffii, a representative lycophyte. It might be expected that auxin has an impact on root development, particularly through meristem bifurcation in dichotomous branching. Surprisingly, the main result of this paper is that exogenous auxin affects root cell elongation but not root cell precursor formation nor branching. Despite auxin increasing root branching in the long term as was shown previously (10.1111/nph.12183), no differences were found in the time needed for the first bifurcation, suggesting that the long-term effect could be indirect. Plants transiently exposed to the hormone or treated with inhibitors are similar to controls, indicating that the early stage of root branching is independent of auxin. This paper is an example of the importance of sharing “negative” results and also points out that auxin’s roles may be not conserved in all plant species in the same way. (Summary by Facundo Romani). Front. Plant Sci. 10.3389/fpls.2019.00154
Auxin is crucial for root development across angiosperms. All the auxin signal transduction pathway components are conserved in land plants, but their role is still not completely clear in early diverged lineages. Here, Fang et al. present a comprehensive analysis of exogenous auxin addition and polar transport inhibitor effects in the root development of Selaginella moellendorffii, a representative lycophyte. It might be expected that auxin has an impact on root development, particularly through meristem bifurcation in dichotomous branching. Surprisingly, the main result of this paper is that exogenous auxin affects root cell elongation but not root cell precursor formation nor branching. Despite auxin increasing root branching in the long term as was shown previously (10.1111/nph.12183), no differences were found in the time needed for the first bifurcation, suggesting that the long-term effect could be indirect. Plants transiently exposed to the hormone or treated with inhibitors are similar to controls, indicating that the early stage of root branching is independent of auxin. This paper is an example of the importance of sharing “negative” results and also points out that auxin’s roles may be not conserved in all plant species in the same way. (Summary by Facundo Romani). Front. Plant Sci. 10.3389/fpls.2019.00154
ABA mediated drought-accelerated flowering ($)
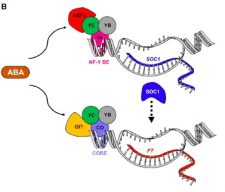
One of the most plastic developmental process in plants is the timing of flower initiation. Plants select favourable conditions to achieve reproductive success. Light and temperature are continuously monitored through molecular mechanisms that are still poorly understood. Drought escape (DE) is an adaptive mechanism which involves rapid flowering to enable the completion of the full life-cycle and seed set in spite of a drought event. In a new paper, Hwang et al. describe a class of abscisic acid (ABA)-related bZIP transcription factors, ABRE-BINDING FACTORS (ABFs), as key drivers for DE. In particular ABA signals are integrated by ABF3/4 into the floral network with the consequent transcriptional activation of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1). At the same time, the photoperiodic pathway is also important for DE: ABA modulates GIGANTEA (GI) signaling and consequently its ability to activate the florigen genes. Take home message: ABA signaling provides different developmental information that allows plants to coordinate the balance between the vegetative and the reproduction phases, according to the available water resources. (Summarized by Francesca Resentini) 10.1016/j.molp.2019.01.002 See also the Spotlight by Conti 10.1016/j.molp.2019.02.002
Convergent gene loss in aquatic plants predicts new components of plant immunity and drought response
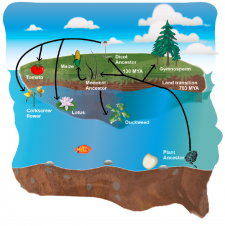 Plants transitioned from water to land around 450 Million years ago. Since their emergence onto land, there have been at least a few reversion events to aquatic environments (e.g., duckweed, humped bladderwort). These big evolutionary events imply adaptation to completely different kinds of biotic and abiotic stresses, requiring many genetic innovations and gene-losses. Here, Baggs et al. studied immune receptors evolution in a wide angiosperm range. Interestingly, they found evidence of convergent gene-loss of particular NLR gene family clades in both monocot and dicot aquatic plant species. They performed genome-wide searches of other genes with a similar evolution pattern, finding another 44 genes recurrently lost in aquatic species. Among them, some are already known for their involvement in biotic and abiotic stress and some are good novel candidates. Gene-loss events are often overlooked in evolutionary literature because they are hard to differentiate from random gene-cleaning processes. This paper highlights their importance and how convergent evolution can help to understand and discover genes associated with major environmental transitions. (Summary by Facundo Romani). bioRxiv 10.1101/572560
Plants transitioned from water to land around 450 Million years ago. Since their emergence onto land, there have been at least a few reversion events to aquatic environments (e.g., duckweed, humped bladderwort). These big evolutionary events imply adaptation to completely different kinds of biotic and abiotic stresses, requiring many genetic innovations and gene-losses. Here, Baggs et al. studied immune receptors evolution in a wide angiosperm range. Interestingly, they found evidence of convergent gene-loss of particular NLR gene family clades in both monocot and dicot aquatic plant species. They performed genome-wide searches of other genes with a similar evolution pattern, finding another 44 genes recurrently lost in aquatic species. Among them, some are already known for their involvement in biotic and abiotic stress and some are good novel candidates. Gene-loss events are often overlooked in evolutionary literature because they are hard to differentiate from random gene-cleaning processes. This paper highlights their importance and how convergent evolution can help to understand and discover genes associated with major environmental transitions. (Summary by Facundo Romani). bioRxiv 10.1101/572560