Plant microbe co-evolution: Allicin resistance in Pseudomonas fluorescens (bioRxiv)
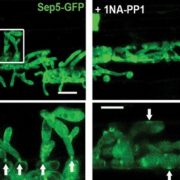
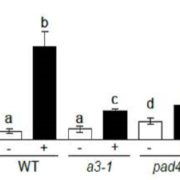
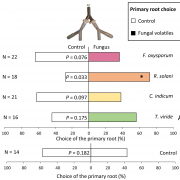
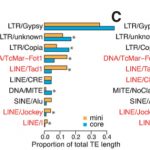
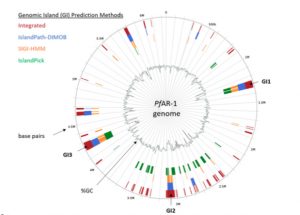 Garlic (Allium sativum L.) produces allicin (diallylthiosulfinate), which is an antibiotic defense substance. It can oxidize thiols in celular targets such as cysteines and glutathione. Because allicin has multiple sites and mechanisms of action, it is difficult for an organism to become resistant. Borlinghaus et al. isolated a highly allicin resistant Pseudomonas fluorescens strain (PfAR-1) from garlic. They used shotgun genomic cloning and whole genome sequencing to functionally identify the genes involved in conferring allicin resistance. Both approaches identified a similar set of genes grouped in three genomic islands; the genes encode mainly redox-related functions. The low GC content and absence of near related sequences to the genomic islands suggests that these regions might have arisen by horizontal gene transfer (HGT). The authors demonstrated the interdependent role of each gene within the genomic islands in creating resistance. Although the origin of these genomic islands remains unknown, related sequences were found in other Pseudomonas species including some that are known plant pathogens. (Summary by Mugdha Sabele) bioRxiv 10.1101/769265
Garlic (Allium sativum L.) produces allicin (diallylthiosulfinate), which is an antibiotic defense substance. It can oxidize thiols in celular targets such as cysteines and glutathione. Because allicin has multiple sites and mechanisms of action, it is difficult for an organism to become resistant. Borlinghaus et al. isolated a highly allicin resistant Pseudomonas fluorescens strain (PfAR-1) from garlic. They used shotgun genomic cloning and whole genome sequencing to functionally identify the genes involved in conferring allicin resistance. Both approaches identified a similar set of genes grouped in three genomic islands; the genes encode mainly redox-related functions. The low GC content and absence of near related sequences to the genomic islands suggests that these regions might have arisen by horizontal gene transfer (HGT). The authors demonstrated the interdependent role of each gene within the genomic islands in creating resistance. Although the origin of these genomic islands remains unknown, related sequences were found in other Pseudomonas species including some that are known plant pathogens. (Summary by Mugdha Sabele) bioRxiv 10.1101/769265