Noriyuki Konishi: The Plant Cell First Author

 Noriyuki Konishi, first author of “Polar localization of a rice silicon transporter requires isoleucine at both C- and N-termini as well as positively charged residues”
Noriyuki Konishi, first author of “Polar localization of a rice silicon transporter requires isoleucine at both C- and N-termini as well as positively charged residues”
Current Position:
Assistant professor, Okayama University, Japan
Education:
B.S. (2008-2012) and Ph.D. (2012-2017) in Graduate School of Agricultural Science, Tohoku University, Japan
Brief bio:
Before starting my academic career, I devoted most of my time to horses because I was a member of the equestrian team at Tohoku University. Honestly, I was not motivated to study science at that time, but after I joined Tomoyuki Yamaya’s lab of plant nutrition, I gradually became interest in studying plant science, similar to being fascinated with my lovely horses. During my Ph.D. course, I investigated the mechanisms of ammonium assimilation in Arabidopsis. After that, I joined Jian Feng Ma’s lab as a post-doctoral researcher in 2017 and started working on the mechanism underlying the polar localization of mineral transporters in rice, especially silicon transporter OsLsi1. In the present study, we identified critical residues required for the polar localization of OsLsi1 and revealed the importance of polar localization in efficient silicon uptake. Since the mechanisms for the polar localization of most mineral transporters are still unclear, especially in rice, I will continue to work on this project to gain the whole picture of polar localization mechanisms.



 Alexander J. Cummins, first author of
Alexander J. Cummins, first author of  Katie M. Murphy, first author of
Katie M. Murphy, first author of  Fabin Yang, co-first author of
Fabin Yang, co-first author of  Jiawei Xiong, co-first author of
Jiawei Xiong, co-first author of  Kai-Chun Peng, first author of
Kai-Chun Peng, first author of 
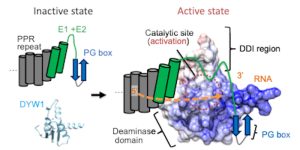 Findings: We determined the crystal structure of the N-terminally truncated DYW domain from Arabidopsis thaliana DYW1. DYW1 forms a cytidine deaminase fold and a C-terminal DYW motif harboring two zinc ions but lacks two structurally conserved β-strands corresponding to the PG box. The recruitment of the PG box and its flanking region from CRR4 likely enables the pocket to accommodate the substrate, conferring catalytic activity to DYW1. In vivo RNA editing assays corroborate that the presence of a PG box in the vicinity of DYW1 is required for its deaminase activity.
Findings: We determined the crystal structure of the N-terminally truncated DYW domain from Arabidopsis thaliana DYW1. DYW1 forms a cytidine deaminase fold and a C-terminal DYW motif harboring two zinc ions but lacks two structurally conserved β-strands corresponding to the PG box. The recruitment of the PG box and its flanking region from CRR4 likely enables the pocket to accommodate the substrate, conferring catalytic activity to DYW1. In vivo RNA editing assays corroborate that the presence of a PG box in the vicinity of DYW1 is required for its deaminase activity.