Perspective: How should the advancement of large language models affect the practice of science?
 In this thought-provoking Perspective, four sets of authors express their opinions about the use of Large Language Models (such as ChatGPT) in the practice of science. Each essay is well reasoned, and each identifies both strengths and limitations of LLMs. There’s agreement that LLMs are good for some things, such as transcribing audio recordings and searching for related manuscripts. However, these applications are vastly different than some of those proposed for LLMs, such as peer review of research publications, or anticipating how humans might respond to a survey. I encourage you to read this article and discuss it with your students or peers, as it is clearly something we all must be informed about. Personally, I fear that widespread use of LLMs will negatively impact the development of critical reading and writing skills of trainees, but I also worry about the perpetuation of inaccuracies as the products of LLMs feed forward into other LLM-crafted texts. Finally, the broad-scale elimination of scientific jobs occurring presently is in part being justified by the ability of LLMs and other AI approaches to “replace” human intellect, an idea that is morally and intellectually repellant. (Summary by Mary Williams @PlantTeaching.bsky.social) PNAS 10.1073/pnas.2401227121
In this thought-provoking Perspective, four sets of authors express their opinions about the use of Large Language Models (such as ChatGPT) in the practice of science. Each essay is well reasoned, and each identifies both strengths and limitations of LLMs. There’s agreement that LLMs are good for some things, such as transcribing audio recordings and searching for related manuscripts. However, these applications are vastly different than some of those proposed for LLMs, such as peer review of research publications, or anticipating how humans might respond to a survey. I encourage you to read this article and discuss it with your students or peers, as it is clearly something we all must be informed about. Personally, I fear that widespread use of LLMs will negatively impact the development of critical reading and writing skills of trainees, but I also worry about the perpetuation of inaccuracies as the products of LLMs feed forward into other LLM-crafted texts. Finally, the broad-scale elimination of scientific jobs occurring presently is in part being justified by the ability of LLMs and other AI approaches to “replace” human intellect, an idea that is morally and intellectually repellant. (Summary by Mary Williams @PlantTeaching.bsky.social) PNAS 10.1073/pnas.2401227121
Apoplastic pH acts as a chemical switch in plants by modulating H2O2 redox potential
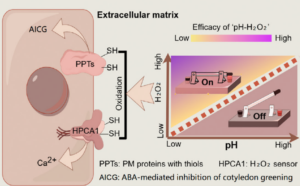 Environmental stimuli, such as drought and salinity, alter cellular conditions including apoplastic pH (pHApo) in plants. These stimuli often lead to an elevation in pHApo which is closely associated with cell functions and plant growth. However, the mechanism behind is largely unexplored. A recent study suggests that pHApo acts as a chemical switch, regulating signalling and modulating stress responses. Using Arabidopsis as the model, Zhou et al. tested the effect of abscisic acid (ABA) on cotyledon greening inhibition under different pHApo. ABA inhibits cotyledon greening by inducing apoplastic H2O2 production. Disrupting apoplastic H2O2 production alleviates the inhibition, and the elevation of pHApo mimics this effect. One might assume that pHApo elevation inhibits apoplastic H2O2 production, but the study reveals a different mechanism: instead of affecting H2O2 levels, increased pHApo lowers the redox potential. Since H2O2 oxidizes cysteine thiols of plasma membrane proteins to transduce stress signals, the lowered redox potential weakens this signal transduction, ultimately leading to stress alleviation. Using the plasma membrane-anchoring calcium signal regulator HPCA1 as an example, the authors showed that elevated pHApo weakened the H2O2-induced oxidation of HPCA1, thereby diminishing calcium signalling, and decreasing the plant sensitivity to H2O2 treatment. The study provides mechanistic insights into pHApo-mediated stress alleviation and highlights the potential to modulate stress responses by manipulating the chemical properties of signalling molecules. (Summary by Yee-Shan Ku @YeeShanKu1) New Phytol. 10.1111/nph.20400
Environmental stimuli, such as drought and salinity, alter cellular conditions including apoplastic pH (pHApo) in plants. These stimuli often lead to an elevation in pHApo which is closely associated with cell functions and plant growth. However, the mechanism behind is largely unexplored. A recent study suggests that pHApo acts as a chemical switch, regulating signalling and modulating stress responses. Using Arabidopsis as the model, Zhou et al. tested the effect of abscisic acid (ABA) on cotyledon greening inhibition under different pHApo. ABA inhibits cotyledon greening by inducing apoplastic H2O2 production. Disrupting apoplastic H2O2 production alleviates the inhibition, and the elevation of pHApo mimics this effect. One might assume that pHApo elevation inhibits apoplastic H2O2 production, but the study reveals a different mechanism: instead of affecting H2O2 levels, increased pHApo lowers the redox potential. Since H2O2 oxidizes cysteine thiols of plasma membrane proteins to transduce stress signals, the lowered redox potential weakens this signal transduction, ultimately leading to stress alleviation. Using the plasma membrane-anchoring calcium signal regulator HPCA1 as an example, the authors showed that elevated pHApo weakened the H2O2-induced oxidation of HPCA1, thereby diminishing calcium signalling, and decreasing the plant sensitivity to H2O2 treatment. The study provides mechanistic insights into pHApo-mediated stress alleviation and highlights the potential to modulate stress responses by manipulating the chemical properties of signalling molecules. (Summary by Yee-Shan Ku @YeeShanKu1) New Phytol. 10.1111/nph.20400
Diversity and genetic basis of hydropatterning in inbred maize lines
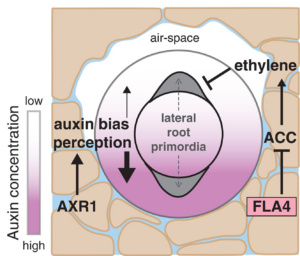 For more than a hundred years, plant biologists have been fascinated by how plants sense and respond to environments that are spatially and temporally heterogenous. Many of these responses occur through the remarkable developmental plasticity of plant growth, such as phototropism and photomorphogenesis. Below ground, root growth and branching patterns as well as root hair formation are highly tuned to the availability of water, nutrients, and even oxygen in the soil matrix. Hydropatterning is the process by which roots respond to water availability through changes in lateral root production. Studies of hydropatterning have been carried out in Arabidopsis, but in this new work, Scharwies et al. determined the extent of hydropatterning in 250 inbred maize lines. The authors found that the percentage of lateral roots forming on the dry size of the root ranged from 0 to 39%; higher proportions of dry-side lateral roots indicates weaker hydropatterning. Interestingly, the inbred lines with stronger hydropatterning also showed deeper roots systems with more brace roots, suggesting that the more efficient placement of lateral roots might allow root resources to be used more efficiently. The authors used GWAS and TWAS approaches to investigate the genetic controls underpinning hydropatterning, and identified several genes involved in auxin signaling, similar to what has been observed in Arabidopsis, as well as genes involved in ethylene synthesis and signaling. The authors also identified a role for an fasciclin-like arabinogalactan protein in hydropatterning, potentially through sensing of environmental cues. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.ads5999
For more than a hundred years, plant biologists have been fascinated by how plants sense and respond to environments that are spatially and temporally heterogenous. Many of these responses occur through the remarkable developmental plasticity of plant growth, such as phototropism and photomorphogenesis. Below ground, root growth and branching patterns as well as root hair formation are highly tuned to the availability of water, nutrients, and even oxygen in the soil matrix. Hydropatterning is the process by which roots respond to water availability through changes in lateral root production. Studies of hydropatterning have been carried out in Arabidopsis, but in this new work, Scharwies et al. determined the extent of hydropatterning in 250 inbred maize lines. The authors found that the percentage of lateral roots forming on the dry size of the root ranged from 0 to 39%; higher proportions of dry-side lateral roots indicates weaker hydropatterning. Interestingly, the inbred lines with stronger hydropatterning also showed deeper roots systems with more brace roots, suggesting that the more efficient placement of lateral roots might allow root resources to be used more efficiently. The authors used GWAS and TWAS approaches to investigate the genetic controls underpinning hydropatterning, and identified several genes involved in auxin signaling, similar to what has been observed in Arabidopsis, as well as genes involved in ethylene synthesis and signaling. The authors also identified a role for an fasciclin-like arabinogalactan protein in hydropatterning, potentially through sensing of environmental cues. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.ads5999
Healing Plants: how bacterial cellulose boosts plant regeneration
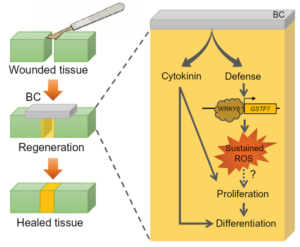 Have you ever placed a leaf from a Monstera plant in a glass of water and watched it grow new roots, forming a new plant? This happens because plants have a much greater ability to regenerate their tissues compared to animals. Recently, in Ruiz-Solaní et al., Spanish researchers have found a way to improve this regeneration, applying bacterial cellulose (BC) to plant wounds. BC is a biopolymer produced by specific bacterial strains that has been widely utilized in human biomedical applications as a scaffold for tissue regeneration. They found that applying bacterial cellulose to artificially cut leaves improved tissue regeneration—in other words, the cuts healed. Additionally, the compound contains plant hormones, such as cytokinin and auxins, which promote cell proliferation and differentiation. Additionally, BC activates plant defense mechanisms against microbial threats, particularly by inducing a reactive oxygen species (ROS) burst, a key response that further supports tissue regeneration. These findings not only shed light on alternative regenerative processes in plants but also open new possibilities for biotechnological applications aimed at improving plant resilience and recovery after injury. (Summary by Carlos González Sanz @carlosgonzsanz.bsky.social) Sci. Advances 10.1126/sciadv.adr1509
Have you ever placed a leaf from a Monstera plant in a glass of water and watched it grow new roots, forming a new plant? This happens because plants have a much greater ability to regenerate their tissues compared to animals. Recently, in Ruiz-Solaní et al., Spanish researchers have found a way to improve this regeneration, applying bacterial cellulose (BC) to plant wounds. BC is a biopolymer produced by specific bacterial strains that has been widely utilized in human biomedical applications as a scaffold for tissue regeneration. They found that applying bacterial cellulose to artificially cut leaves improved tissue regeneration—in other words, the cuts healed. Additionally, the compound contains plant hormones, such as cytokinin and auxins, which promote cell proliferation and differentiation. Additionally, BC activates plant defense mechanisms against microbial threats, particularly by inducing a reactive oxygen species (ROS) burst, a key response that further supports tissue regeneration. These findings not only shed light on alternative regenerative processes in plants but also open new possibilities for biotechnological applications aimed at improving plant resilience and recovery after injury. (Summary by Carlos González Sanz @carlosgonzsanz.bsky.social) Sci. Advances 10.1126/sciadv.adr1509
ATP-citrate lyase B1 regulates pollen development in Arabidopsis
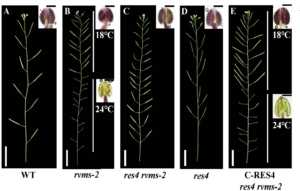 The genetic regulation of male fertility in Arabidopsis thaliana involves multiple factors, including ATP-Citrate Lyase (ACL), a key enzyme in cytoplasmic acetyl-CoA production. The thermosensitive male-sterile mutant reversible male sterile-2 (rvms-2) exhibits normal fertility at low temperatures but becomes male sterile at higher temperatures. This mutant shows defective exine formation and cytoplasmic leakage during pollen development. RVMS encodes a GDSL lipase/hydrolase protein with lipase activity, essential for proper pollen wall formation. Previous studies identified several rvms-2 suppressor mutants that restore fertility. Among these, restorer-2 (res2) and restorer-3 (res3) encode cell-wall-modifying enzymes that delay degradation of the pectin and callose walls, thereby improving pollen viability. In this new study, Ma et al. characterized restorer-4 (res4), another suppressor mutant of rvms-2, revealing that RES4 encodes ATP-Citrate Lyase B1 (ACLB1), a subunit of ACL that catalyzes the conversion of citrate to acetyl-CoA. The res4 mutation alters the citryl-CoA lyase domain of ACLB1, leading to defective exine formation and reduced microspore size. RES4 is expressed in the tapetum and regulated by the transcription factor MS188, a key player in sporopollenin synthesis. Lipid staining assays indicate significantly reduced lipid accumulation in the tapetum and locules of res4, linking ACL activity to sporopollenin production. The smaller microspore size in res4 contributes to partial restoration of fertility in rvms-2, suggesting that reduced microspore volume lowers the requirement for protective cell wall structures. This study underscores the importance of ACL-derived cytoplasmic acetyl-CoA in pollen development and offers new insights into fertility restoration mechanisms, with potential implications for hybrid crop breeding strategies. (Summary by Muhammad Aamir Khan @MAKNature1998) Plant Physiol. 10.1093/plphys/kiaf044
The genetic regulation of male fertility in Arabidopsis thaliana involves multiple factors, including ATP-Citrate Lyase (ACL), a key enzyme in cytoplasmic acetyl-CoA production. The thermosensitive male-sterile mutant reversible male sterile-2 (rvms-2) exhibits normal fertility at low temperatures but becomes male sterile at higher temperatures. This mutant shows defective exine formation and cytoplasmic leakage during pollen development. RVMS encodes a GDSL lipase/hydrolase protein with lipase activity, essential for proper pollen wall formation. Previous studies identified several rvms-2 suppressor mutants that restore fertility. Among these, restorer-2 (res2) and restorer-3 (res3) encode cell-wall-modifying enzymes that delay degradation of the pectin and callose walls, thereby improving pollen viability. In this new study, Ma et al. characterized restorer-4 (res4), another suppressor mutant of rvms-2, revealing that RES4 encodes ATP-Citrate Lyase B1 (ACLB1), a subunit of ACL that catalyzes the conversion of citrate to acetyl-CoA. The res4 mutation alters the citryl-CoA lyase domain of ACLB1, leading to defective exine formation and reduced microspore size. RES4 is expressed in the tapetum and regulated by the transcription factor MS188, a key player in sporopollenin synthesis. Lipid staining assays indicate significantly reduced lipid accumulation in the tapetum and locules of res4, linking ACL activity to sporopollenin production. The smaller microspore size in res4 contributes to partial restoration of fertility in rvms-2, suggesting that reduced microspore volume lowers the requirement for protective cell wall structures. This study underscores the importance of ACL-derived cytoplasmic acetyl-CoA in pollen development and offers new insights into fertility restoration mechanisms, with potential implications for hybrid crop breeding strategies. (Summary by Muhammad Aamir Khan @MAKNature1998) Plant Physiol. 10.1093/plphys/kiaf044
Symbiotic secrets: A multi-omics exploration of the lichen Xanthoria parietina
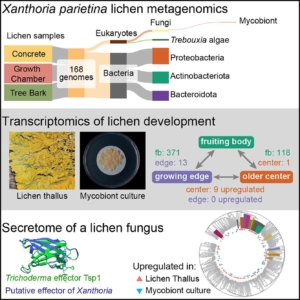 Lichens are complex symbiotic organisms formed by algae or cyanobacteria living within the filaments of multiple fungal species. This unique partnership, which is both stable and self-replicating across generations, represents one of the most successful lifestyle strategies in the biological world. Lichens play a crucial role in nutrient cycling and ecosystem stability, often thriving in extreme environments where few other organisms can survive. Interestingly, lichens exhibit properties distinct from those of their individual components, yet the molecular mechanisms underlying their formation and persistence remain largely unknown. To address this knowledge gap, Tagirdzhanova and colleagues employed metagenomics and metatranscriptomics to investigate the symbiosis in the model lichen Xanthoria parietina. Their study generated a comprehensive dataset encompassing the genome, transcriptome, and secretome, providing a valuable resource for future research on lichen developmental biology. Notably, they identified an enrichment of NLR-like genes and effector proteins in the mycobiont, suggesting that lichen symbiosis involves bidirectional recognition between symbionts and potentially the selective recruitment of compatible microbial partners. (Summary by Ching Chan @ntnuchanlab) Current Biology 10.1016/j.cub.2024.12.041
Lichens are complex symbiotic organisms formed by algae or cyanobacteria living within the filaments of multiple fungal species. This unique partnership, which is both stable and self-replicating across generations, represents one of the most successful lifestyle strategies in the biological world. Lichens play a crucial role in nutrient cycling and ecosystem stability, often thriving in extreme environments where few other organisms can survive. Interestingly, lichens exhibit properties distinct from those of their individual components, yet the molecular mechanisms underlying their formation and persistence remain largely unknown. To address this knowledge gap, Tagirdzhanova and colleagues employed metagenomics and metatranscriptomics to investigate the symbiosis in the model lichen Xanthoria parietina. Their study generated a comprehensive dataset encompassing the genome, transcriptome, and secretome, providing a valuable resource for future research on lichen developmental biology. Notably, they identified an enrichment of NLR-like genes and effector proteins in the mycobiont, suggesting that lichen symbiosis involves bidirectional recognition between symbionts and potentially the selective recruitment of compatible microbial partners. (Summary by Ching Chan @ntnuchanlab) Current Biology 10.1016/j.cub.2024.12.041
Finding balance: How plants achieve signal specificity in stomatal development and defense
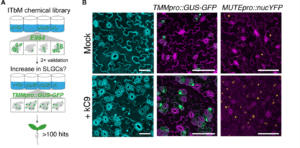 Plants need to develop and grow within their environment whilst defending themselves against potential external threats. There are limited resources for these energy-intensive processes and so cross-talk is common between the signalling pathways to find balance between growth and defence. Hermann et al., provide evidence for cross-regulation between stomatal development and the immune response and suggest that that the key to signal specificity is the balance of activation of signal receptors. Through conducting an intensive chemical screen, Hermann et al. pin-pointed a candidate signalling chemical (kC9) which activates the differentiation of stomatal cells by inhibiting the EPF-ERECTA signalling pathway. Docking models, interferometry and calorimetry assays showed that kC9 binds to the transducers of the signalling cascade, the mitogen-activated protein kinases (MAPKs), specifically MPK3 and MPK6, which inhibit stomatal development. MPK3 and MPK6 are also downstream of the FLS2-flg22 signalling pathway, a well-documented pathway in the plant immune response. flg22 treatment negated the proliferative effect of kC9, as well as the loss of ERECTA family transporters, on stomatal development, demonstrating cross-regulation from the immune pathway. This research highlights the importance of the availability of upstream receptors for signal specificity where there are overlapping components of the pathways. (Summary by Kes Maio @kesmaio.bsky.social) Science Advances 10.1126/sciadv.ads3718
Plants need to develop and grow within their environment whilst defending themselves against potential external threats. There are limited resources for these energy-intensive processes and so cross-talk is common between the signalling pathways to find balance between growth and defence. Hermann et al., provide evidence for cross-regulation between stomatal development and the immune response and suggest that that the key to signal specificity is the balance of activation of signal receptors. Through conducting an intensive chemical screen, Hermann et al. pin-pointed a candidate signalling chemical (kC9) which activates the differentiation of stomatal cells by inhibiting the EPF-ERECTA signalling pathway. Docking models, interferometry and calorimetry assays showed that kC9 binds to the transducers of the signalling cascade, the mitogen-activated protein kinases (MAPKs), specifically MPK3 and MPK6, which inhibit stomatal development. MPK3 and MPK6 are also downstream of the FLS2-flg22 signalling pathway, a well-documented pathway in the plant immune response. flg22 treatment negated the proliferative effect of kC9, as well as the loss of ERECTA family transporters, on stomatal development, demonstrating cross-regulation from the immune pathway. This research highlights the importance of the availability of upstream receptors for signal specificity where there are overlapping components of the pathways. (Summary by Kes Maio @kesmaio.bsky.social) Science Advances 10.1126/sciadv.ads3718
Tomato PR1 protein prevents the fungal effector FolSvp2 from suppressing SlISP-mediated ROS production
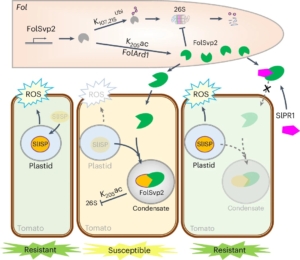 Plants and pathogens are locked in a co-evolutionary arms race. Pathogens secrete effectors into plants, leading to effector-triggered susceptibility. Plants in turn respond through the production of reactive oxygen species (ROS) and the expression of defence-related genes to defend themselves, which leads to effector-triggered immunity. Recently, Li et al. showed that FolSvp2, an effector secreted from the fungal pathogen Fusarium oxysporom f. sp. Lycopersici (Fol) undergoes modification through acetylation. This modification stabilizes this effector through phase separation inside the host cell. Furthermore, the stabilized FolSvp2 recruits into the effector condensates a tomato iron-sulfur protein, SlISP, which is normally plastid-localized and contributes to the defensive ROS burst. This interaction with FolSvp2 prevents SlISP from producing ROS, much like a thief (FolSvp2) holds a householder (SlISP) hostage and prevents it from shouting for help. The authors further showed how SlPR1, a resistance protein in tomato, prevents the condensation of SlISP by interacting with FolSvp2 in the apoplast (like the police blocking a thief from entering the household). Thus, SlPR1 prevents FolSvp2 from sequestering and inactivating SlISP, which then allows defensive ROS production. Their results show a clear example of how pathogens can use non-enzymatic approach to suppress host defenses, and that pathogen effectors also use phase separation, forming condensates within host cells during infection. (Summary by Nathaniel Oragbon @NathanIgwe) Nature Plants 10.1038/s41477-024-01811-y
Plants and pathogens are locked in a co-evolutionary arms race. Pathogens secrete effectors into plants, leading to effector-triggered susceptibility. Plants in turn respond through the production of reactive oxygen species (ROS) and the expression of defence-related genes to defend themselves, which leads to effector-triggered immunity. Recently, Li et al. showed that FolSvp2, an effector secreted from the fungal pathogen Fusarium oxysporom f. sp. Lycopersici (Fol) undergoes modification through acetylation. This modification stabilizes this effector through phase separation inside the host cell. Furthermore, the stabilized FolSvp2 recruits into the effector condensates a tomato iron-sulfur protein, SlISP, which is normally plastid-localized and contributes to the defensive ROS burst. This interaction with FolSvp2 prevents SlISP from producing ROS, much like a thief (FolSvp2) holds a householder (SlISP) hostage and prevents it from shouting for help. The authors further showed how SlPR1, a resistance protein in tomato, prevents the condensation of SlISP by interacting with FolSvp2 in the apoplast (like the police blocking a thief from entering the household). Thus, SlPR1 prevents FolSvp2 from sequestering and inactivating SlISP, which then allows defensive ROS production. Their results show a clear example of how pathogens can use non-enzymatic approach to suppress host defenses, and that pathogen effectors also use phase separation, forming condensates within host cells during infection. (Summary by Nathaniel Oragbon @NathanIgwe) Nature Plants 10.1038/s41477-024-01811-y
Shaping pathogenicity: How CRISPR-Cas loss fuels Xanthomonas evolution
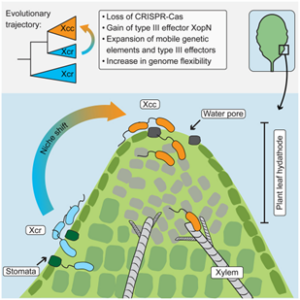 Pathogens have evolved diverse infection strategies governed by virulence factors, often targeting specific host organs or tissues. Genome fluidity plays a crucial role in enabling microbial pathogens to adapt to the dynamic selection pressures imposed by co-evolution with their hosts. In a recent study, Paauw and colleagues used a pangenome approach to investigate the genetic factors and evolutionary processes shaping the evolution of pathogenic Xanthomonas campestris (Xc) in a collection of over 180 isolates from diseased Brassica plants. Notably, different Xc pathovars exhibit distinct tissue specificities within the same leaf: Xc pv. campestris (Xcc) infects the vasculature, whereas Xc pv. raphanin (Xcr) enters leaves exclusively through stomata, like most bacteria. However, the genetic mechanisms driving the diversification of virulence factors remain unclear. Prokaryotic CRISPR-Cas systems serve as adaptive immune defenses that restrict horizontal gene transfer and limit the uptake of foreign DNA. The authors found that loss of CRISPR-Cas is more prominent in the Xcc genome compared to Xcr, facilitating the acquisition of virulence factors, and increasing genome plasticity. This enhanced genomic flexibility allows Xcc to colonize additional plant tissues that other pathovars cannot. While gene regulations can occur rapidly during host infection, the emergence of new species or subspecies requires continuous genome evolution. Overall, these findings suggest that CRISPR-Cas loss has been a key driver of Xc evolution, contributing to the emergence of agronomically important vascular pathogens in cabbage. (Summary by Ching Chan @ntnuchanlab) Curr. Biol. 10.1016/j.cub.2025.01.003
Pathogens have evolved diverse infection strategies governed by virulence factors, often targeting specific host organs or tissues. Genome fluidity plays a crucial role in enabling microbial pathogens to adapt to the dynamic selection pressures imposed by co-evolution with their hosts. In a recent study, Paauw and colleagues used a pangenome approach to investigate the genetic factors and evolutionary processes shaping the evolution of pathogenic Xanthomonas campestris (Xc) in a collection of over 180 isolates from diseased Brassica plants. Notably, different Xc pathovars exhibit distinct tissue specificities within the same leaf: Xc pv. campestris (Xcc) infects the vasculature, whereas Xc pv. raphanin (Xcr) enters leaves exclusively through stomata, like most bacteria. However, the genetic mechanisms driving the diversification of virulence factors remain unclear. Prokaryotic CRISPR-Cas systems serve as adaptive immune defenses that restrict horizontal gene transfer and limit the uptake of foreign DNA. The authors found that loss of CRISPR-Cas is more prominent in the Xcc genome compared to Xcr, facilitating the acquisition of virulence factors, and increasing genome plasticity. This enhanced genomic flexibility allows Xcc to colonize additional plant tissues that other pathovars cannot. While gene regulations can occur rapidly during host infection, the emergence of new species or subspecies requires continuous genome evolution. Overall, these findings suggest that CRISPR-Cas loss has been a key driver of Xc evolution, contributing to the emergence of agronomically important vascular pathogens in cabbage. (Summary by Ching Chan @ntnuchanlab) Curr. Biol. 10.1016/j.cub.2025.01.003
LecRK-V and trehalose-associated immunity: a conserved defense mechanism across kingdoms
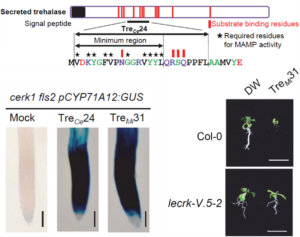 Membrane-localized receptor-like kinases (RLKs) are essential for perceiving diverse signaling molecules, including proteins, polysaccharides, hormones, reactive oxygen species, ions, and damage-associated molecular patterns. Among the well-characterized RLKs, FLS2, EFR, and CERK1 are known to recognize bacterial and fungal molecular patterns, triggering plant immune responses. However, the Arabidopsis genome encodes approximately 600 RLKs, the majority of which remain functionally uncharacterized. In a recent study, Iino, Kadota, and colleagues investigated how Arabidopsis perceives nematodes, uncovering a trehalase-derived peptide that activates immune responses in the Col-0 ecotype but not in Cvi-0. Through genetic crosses and QTL mapping, the authors identified members of the LecRK-V family as receptors for this peptide. Intriguingly, this ligand-receptor pair is evolutionarily conserved across insects and fungal pathogens, underscoring the broad-spectrum role in plant defense. This study highlights the critical function of the LecRK-V family in pathogen perception and suggests that trehalase/trehalose-associated components serve as conserved immune signals across different biological kingdoms. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2025.02.02.636166
Membrane-localized receptor-like kinases (RLKs) are essential for perceiving diverse signaling molecules, including proteins, polysaccharides, hormones, reactive oxygen species, ions, and damage-associated molecular patterns. Among the well-characterized RLKs, FLS2, EFR, and CERK1 are known to recognize bacterial and fungal molecular patterns, triggering plant immune responses. However, the Arabidopsis genome encodes approximately 600 RLKs, the majority of which remain functionally uncharacterized. In a recent study, Iino, Kadota, and colleagues investigated how Arabidopsis perceives nematodes, uncovering a trehalase-derived peptide that activates immune responses in the Col-0 ecotype but not in Cvi-0. Through genetic crosses and QTL mapping, the authors identified members of the LecRK-V family as receptors for this peptide. Intriguingly, this ligand-receptor pair is evolutionarily conserved across insects and fungal pathogens, underscoring the broad-spectrum role in plant defense. This study highlights the critical function of the LecRK-V family in pathogen perception and suggests that trehalase/trehalose-associated components serve as conserved immune signals across different biological kingdoms. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2025.02.02.636166
 Membrane-localized receptor-like kinases (RLKs) are essential for perceiving diverse signaling molecules, including proteins, polysaccharides, hormones, reactive oxygen species, ions, and damage-associated molecular patterns. Among the well-characterized RLKs, FLS2, EFR, and CERK1 are known to recognize bacterial and fungal molecular patterns, triggering plant immune responses. However, the Arabidopsis genome encodes approximately 600 RLKs, the majority of which remain functionally uncharacterized. In a recent study, Iino, Kadota, and colleagues investigated how Arabidopsis perceives nematodes, uncovering a trehalase-derived peptide that activates immune responses in the Col-0 ecotype but not in Cvi-0. Through genetic crosses and QTL mapping, the authors identified members of the LecRK-V family as receptors for this peptide. Intriguingly, this ligand-receptor pair is evolutionarily conserved across insects and fungal pathogens, underscoring the broad-spectrum role in plant defense. This study highlights the critical function of the LecRK-V family in pathogen perception and suggests that trehalase/trehalose-associated components serve as conserved immune signals across different biological kingdoms. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2025.02.02.636166
Membrane-localized receptor-like kinases (RLKs) are essential for perceiving diverse signaling molecules, including proteins, polysaccharides, hormones, reactive oxygen species, ions, and damage-associated molecular patterns. Among the well-characterized RLKs, FLS2, EFR, and CERK1 are known to recognize bacterial and fungal molecular patterns, triggering plant immune responses. However, the Arabidopsis genome encodes approximately 600 RLKs, the majority of which remain functionally uncharacterized. In a recent study, Iino, Kadota, and colleagues investigated how Arabidopsis perceives nematodes, uncovering a trehalase-derived peptide that activates immune responses in the Col-0 ecotype but not in Cvi-0. Through genetic crosses and QTL mapping, the authors identified members of the LecRK-V family as receptors for this peptide. Intriguingly, this ligand-receptor pair is evolutionarily conserved across insects and fungal pathogens, underscoring the broad-spectrum role in plant defense. This study highlights the critical function of the LecRK-V family in pathogen perception and suggests that trehalase/trehalose-associated components serve as conserved immune signals across different biological kingdoms. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2025.02.02.636166










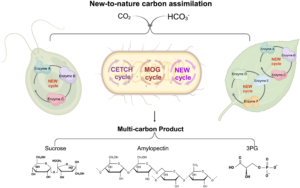 Rubisco (Ribulose‐1,5‐bisphosphate carboxylase/oxygenase) is the central enzyme for photosynthesis, This enzyme poorly discriminates between CO2 and O2, which limits its efficiency. To work around this and make carbon assimilation more efficient, scientists have been employing different engineering strategies, several of which are summarized in a new review by Qin et al. One strategy is to alter Rubisco activity, for example by inducing mutations, combining parts of Rubisco from different organisms, engineering other enzymes involved with Rubisco activation and accumulation, and introducing Rubisco genes from lower organisms to crop plants. Another engineering approach is to create carbon-concentrating compartments, for example by using carboxysomes found in cyanobacteria and pyrenoids from eukaryotic algae. Alternatively, CO2 levels at Rubisco can be boosted by introducing the C4 cycle in C3 plants. Scientists are also engineering the Calvin-Benson-Bassham Cycle by overexpressing specific enzymes involved in the cycle, as well as adding photorespiratory bypasses to reduce carbon loss due to photorespiration. Lastly, work has been done to design a new artificial carbon assimilation cycle by using deep learning and synthetic biology. Synthetic biology approaches can also aid in developing artificial enzyme complexes that could improve photosynthesis in plants. (Summary by Mae Mercado
Rubisco (Ribulose‐1,5‐bisphosphate carboxylase/oxygenase) is the central enzyme for photosynthesis, This enzyme poorly discriminates between CO2 and O2, which limits its efficiency. To work around this and make carbon assimilation more efficient, scientists have been employing different engineering strategies, several of which are summarized in a new review by Qin et al. One strategy is to alter Rubisco activity, for example by inducing mutations, combining parts of Rubisco from different organisms, engineering other enzymes involved with Rubisco activation and accumulation, and introducing Rubisco genes from lower organisms to crop plants. Another engineering approach is to create carbon-concentrating compartments, for example by using carboxysomes found in cyanobacteria and pyrenoids from eukaryotic algae. Alternatively, CO2 levels at Rubisco can be boosted by introducing the C4 cycle in C3 plants. Scientists are also engineering the Calvin-Benson-Bassham Cycle by overexpressing specific enzymes involved in the cycle, as well as adding photorespiratory bypasses to reduce carbon loss due to photorespiration. Lastly, work has been done to design a new artificial carbon assimilation cycle by using deep learning and synthetic biology. Synthetic biology approaches can also aid in developing artificial enzyme complexes that could improve photosynthesis in plants. (Summary by Mae Mercado 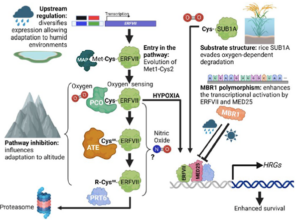 Hypoxia, or reduced oxygen availability, is a double-edged sword: while it disrupts metabolism and can cause cell death, it also plays a vital role in regulating development in animals and plants. With extreme flooding increasing due to climate change, understanding how genetic variation enhances hypoxia tolerance provides valuable strategies for improving crop resilience. This review by Holdsworth et al. examines how geography, altitude, and agriculture have shaped molecular responses to hypoxia, particularly through the PLANT CYSTEINE OXIDASE (PCO) N-degron pathway, and how these responses have evolved across different land plant species over time. Local adaptation plays a key role in acclimatization to humid environments, where flooding, often triggered by intense rainfall, drives acute hypoxic stress. Environmental genome-wide association studies (eGWAS) help identify genes linked to hypoxia tolerance by integrating large-scale genomic and environmental data. Altitude also influences oxygen-sensing adaptations, though no direct molecular mechanism for absolute altitude sensing has been identified. However, adaptation to altitude is at least partially linked to the PCO N-degron pathway. Agriculture has further shaped hypoxia responses, as seen in rice: the FR13A variety uses the Sub1A-1 gene to withstand full submersion by limiting growth and enhancing anaerobic metabolism, while deepwater rice employs SNORKEL1/2 genes to elongate stems and escape partial submersion. These insights drive breeding strategies for flood-resistant crops. Ultimately, understanding the conserved PCO N-degron pathway provides a foundation for future research into plant adaptation to low-oxygen environments, integrating georeferenced genomic data and biochemical findings. This article is part of the Plant Physiology focus issue on Hypoxia and Plants; a recorded webinar on this topic
Hypoxia, or reduced oxygen availability, is a double-edged sword: while it disrupts metabolism and can cause cell death, it also plays a vital role in regulating development in animals and plants. With extreme flooding increasing due to climate change, understanding how genetic variation enhances hypoxia tolerance provides valuable strategies for improving crop resilience. This review by Holdsworth et al. examines how geography, altitude, and agriculture have shaped molecular responses to hypoxia, particularly through the PLANT CYSTEINE OXIDASE (PCO) N-degron pathway, and how these responses have evolved across different land plant species over time. Local adaptation plays a key role in acclimatization to humid environments, where flooding, often triggered by intense rainfall, drives acute hypoxic stress. Environmental genome-wide association studies (eGWAS) help identify genes linked to hypoxia tolerance by integrating large-scale genomic and environmental data. Altitude also influences oxygen-sensing adaptations, though no direct molecular mechanism for absolute altitude sensing has been identified. However, adaptation to altitude is at least partially linked to the PCO N-degron pathway. Agriculture has further shaped hypoxia responses, as seen in rice: the FR13A variety uses the Sub1A-1 gene to withstand full submersion by limiting growth and enhancing anaerobic metabolism, while deepwater rice employs SNORKEL1/2 genes to elongate stems and escape partial submersion. These insights drive breeding strategies for flood-resistant crops. Ultimately, understanding the conserved PCO N-degron pathway provides a foundation for future research into plant adaptation to low-oxygen environments, integrating georeferenced genomic data and biochemical findings. This article is part of the Plant Physiology focus issue on Hypoxia and Plants; a recorded webinar on this topic 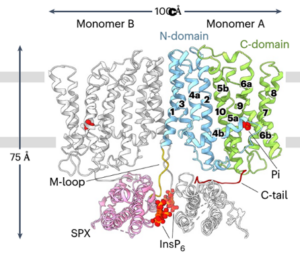 Phosphorus (P) is an essential macronutrient required for plant growth, development, and reproduction. It is primarily absorbed by plant roots in the form of orthophosphate (Pi). The root-to-shoot translocation of Pi depends on a crucial xylem-loading process mediated by PHOSPHATE 1 (PHO1), a Pi efflux transporter expressed in the pericycle and xylem parenchyma cells. Mutations in PHO1 result in severely impaired shoot growth and reduced seed production. Although the physiological role of PHO1 has been recognized for over two decades, its structure and gating mechanism were only recently elucidated. Using a combination of crystallography and modeling tools, Fang and colleagues resolved the Arabidopsis PHO1;H1 (AtPHO1;H1) structure, revealing an N-terminal cytoplasmic SPX domain, a transmembrane EXS domain, and a partially disordered C-terminal tail. The EXS domain facilitates Pi translocation and is gated by the residues Trp719 and Tyr610. The SPX domain binds PP-InsP molecules to regulate transport activity, while the C-terminal tail contributes to dimer formation and PP-InsP binding. These findings provide critical insights for optimizing phosphorus-use efficiency (PUE) in plants, which is essential for reducing dependence on P fertilizers and promoting sustainable, environmentally friendly agriculture. (Summary by Ching Chan
Phosphorus (P) is an essential macronutrient required for plant growth, development, and reproduction. It is primarily absorbed by plant roots in the form of orthophosphate (Pi). The root-to-shoot translocation of Pi depends on a crucial xylem-loading process mediated by PHOSPHATE 1 (PHO1), a Pi efflux transporter expressed in the pericycle and xylem parenchyma cells. Mutations in PHO1 result in severely impaired shoot growth and reduced seed production. Although the physiological role of PHO1 has been recognized for over two decades, its structure and gating mechanism were only recently elucidated. Using a combination of crystallography and modeling tools, Fang and colleagues resolved the Arabidopsis PHO1;H1 (AtPHO1;H1) structure, revealing an N-terminal cytoplasmic SPX domain, a transmembrane EXS domain, and a partially disordered C-terminal tail. The EXS domain facilitates Pi translocation and is gated by the residues Trp719 and Tyr610. The SPX domain binds PP-InsP molecules to regulate transport activity, while the C-terminal tail contributes to dimer formation and PP-InsP binding. These findings provide critical insights for optimizing phosphorus-use efficiency (PUE) in plants, which is essential for reducing dependence on P fertilizers and promoting sustainable, environmentally friendly agriculture. (Summary by Ching Chan 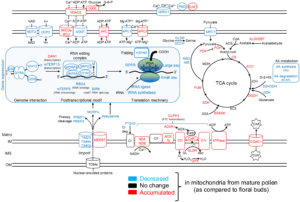 Imagine you’re on a quest to deliver a package, racing against the competition. How do you prepare? Pollen grains and the pollen tubes that they form are essentially package-delivery systems. Their purpose is to deliver genetic information (sperm cell nuclei) to the ovule. Once the task is completed, the pollen cell itself becomes obsolete. A new study by Boussardon et al. investigated the proteome of mature pollen grains which they isolated using the IMTACT protocol: Isolation of Mitochondria Tagged in specific Cell-Type. The authors showed that pollen grain mitochondria switch into a focused energy-generating program, with proteins involved in energy production (TCA cycle and electron transport chain) highly enriched, not unlike a runner carbo-loading before a race. At the same time, proteins involved in transcription and translation are strongly depleted, and the mitochondrial genome itself is actively degraded during maturation; in essence the mitochondria terminally differentiate and channel their resources into their delivery task. This observation is fascinating in light of the fact that the progeny of sexual reproduction inherit their mitochondria solely from the egg-donating parent, and not the sperm-donating parent, no pollen grain mitochondria need to be viable for transfer into the zygote. (Summary by Mary Williams
Imagine you’re on a quest to deliver a package, racing against the competition. How do you prepare? Pollen grains and the pollen tubes that they form are essentially package-delivery systems. Their purpose is to deliver genetic information (sperm cell nuclei) to the ovule. Once the task is completed, the pollen cell itself becomes obsolete. A new study by Boussardon et al. investigated the proteome of mature pollen grains which they isolated using the IMTACT protocol: Isolation of Mitochondria Tagged in specific Cell-Type. The authors showed that pollen grain mitochondria switch into a focused energy-generating program, with proteins involved in energy production (TCA cycle and electron transport chain) highly enriched, not unlike a runner carbo-loading before a race. At the same time, proteins involved in transcription and translation are strongly depleted, and the mitochondrial genome itself is actively degraded during maturation; in essence the mitochondria terminally differentiate and channel their resources into their delivery task. This observation is fascinating in light of the fact that the progeny of sexual reproduction inherit their mitochondria solely from the egg-donating parent, and not the sperm-donating parent, no pollen grain mitochondria need to be viable for transfer into the zygote. (Summary by Mary Williams 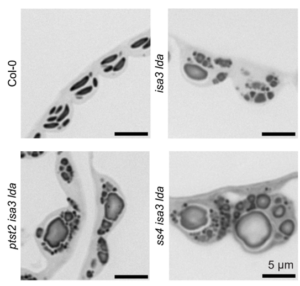 Starch is the major storage carbohydrate in plants, and in Arabidopsis leaves it forms granules in the chloroplasts to supply energy during the night when photosynthesis is inactive. These semicrystalline granules are made of glucose chains, which can be mostly unbranched (amylose) or highly branched (amylopectin). The formation of new starch granules (initiation) is achieved by building on short glucose chains called malto-oligosaccharide (MOS) primers. A key step in this process is the delivery of a glucosyltransferase STARCH SYNTHASE 4 (SS4) to the new granule by PROTEIN TARGETING TO STARCH 2 (PTST2). However, new evidence from Heutinck et al. supports an alternative route for starch granule initiation. In an Arabidopsis mutant deficient in two debranching enzymes, branched MOS in the chloroplast accumulate to levels ten times higher than in the wild type. Additional granules were also formed in these chloroplasts, which were smaller, differently shaped, and not reliant on SS4 or PTST2 to be initiated. While additional research is needed to elucidate the mechanism, the possibility of a new pathway for starch granule initiation provides huge potential for better understanding plant carbon metabolism and hence crop improvement. (Summary by Ciara O’Brien) Plant Physiol.
Starch is the major storage carbohydrate in plants, and in Arabidopsis leaves it forms granules in the chloroplasts to supply energy during the night when photosynthesis is inactive. These semicrystalline granules are made of glucose chains, which can be mostly unbranched (amylose) or highly branched (amylopectin). The formation of new starch granules (initiation) is achieved by building on short glucose chains called malto-oligosaccharide (MOS) primers. A key step in this process is the delivery of a glucosyltransferase STARCH SYNTHASE 4 (SS4) to the new granule by PROTEIN TARGETING TO STARCH 2 (PTST2). However, new evidence from Heutinck et al. supports an alternative route for starch granule initiation. In an Arabidopsis mutant deficient in two debranching enzymes, branched MOS in the chloroplast accumulate to levels ten times higher than in the wild type. Additional granules were also formed in these chloroplasts, which were smaller, differently shaped, and not reliant on SS4 or PTST2 to be initiated. While additional research is needed to elucidate the mechanism, the possibility of a new pathway for starch granule initiation provides huge potential for better understanding plant carbon metabolism and hence crop improvement. (Summary by Ciara O’Brien) Plant Physiol. 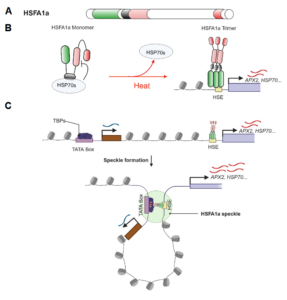 The heat shock response, a rapid transcriptional response to heat, was first observed nearly 60 years ago, and has long been a paradigm for understanding gene responses to exogenous cues. The family of genes encoding heat shock factors (HSFs) is greatly expanded in plants. These HSFs serve as key regulators of the heat shock response, both initiating short-term transcriptional changes, but also serving as a molecular memory that activates heat memory genes after an earlier “priming” stress. A new study by Peng et al. examined how conformational changes in HSF protein structure contributes to both short term and memory responses to heat shock. The authors identified a transcriptional cascade, led by rapid activation HSFA1 by protein conformational changes. Within moments of heat shock, HSFA1 factors form speckles in the nucleus, suggesting that they act as heat sensors, and eliciting a very rapid transcriptional response. HSFA1 factors contain two Prion-related Domains (PrDs), which the authors showed are critical for their thermal responsiveness. PrDs are intrinsically disordered domains that are often found in proteins that undergo liquid phase separation. One of these, PrD1, is responsible for sequestering the HSFA1 proteins during ambient temperatures and maintaining them in an inactive state. The other, PrD2, is required for heat activation and thermal memory, through the formation of DNA loops that connect heat-responsive promoters with enhancer domains. This work builds on the growing recognition of the importance of intrinsically disordered domains and liquid phase separation in modulating rapid cellular responses. (Summary by Mary Williams @PlantTeaching.bsky.social) Mol Plant
The heat shock response, a rapid transcriptional response to heat, was first observed nearly 60 years ago, and has long been a paradigm for understanding gene responses to exogenous cues. The family of genes encoding heat shock factors (HSFs) is greatly expanded in plants. These HSFs serve as key regulators of the heat shock response, both initiating short-term transcriptional changes, but also serving as a molecular memory that activates heat memory genes after an earlier “priming” stress. A new study by Peng et al. examined how conformational changes in HSF protein structure contributes to both short term and memory responses to heat shock. The authors identified a transcriptional cascade, led by rapid activation HSFA1 by protein conformational changes. Within moments of heat shock, HSFA1 factors form speckles in the nucleus, suggesting that they act as heat sensors, and eliciting a very rapid transcriptional response. HSFA1 factors contain two Prion-related Domains (PrDs), which the authors showed are critical for their thermal responsiveness. PrDs are intrinsically disordered domains that are often found in proteins that undergo liquid phase separation. One of these, PrD1, is responsible for sequestering the HSFA1 proteins during ambient temperatures and maintaining them in an inactive state. The other, PrD2, is required for heat activation and thermal memory, through the formation of DNA loops that connect heat-responsive promoters with enhancer domains. This work builds on the growing recognition of the importance of intrinsically disordered domains and liquid phase separation in modulating rapid cellular responses. (Summary by Mary Williams @PlantTeaching.bsky.social) Mol Plant 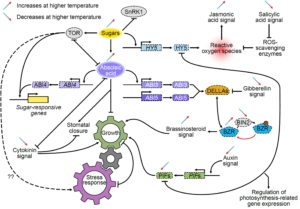 Due to anthropomorphic global warming, 2024 was the first year during which the global mean temperature was more than 1.5° above pre-industrial levels. Clearly, understanding how high temperatures affect plant physiology is an urgent priority. In this new study, Zhang et al. did a multi-parameter study of the model C4 plant Setaria viridis. They grew two populations for two weeks at 28°, and then for the next two weeks one group was grown at 42° whilst the other remained at 28°. After this, the authors measured photosynthesis parameters, and carried out metabolomic, transcriptomic, and proteomic studies, leading to several key findings. As expected, the heat-grown plants were significantly smaller than their counterparts. Surprisingly, their photosynthetic efficiency was hardly altered. The heat-grown plants had much higher levels of hexoses, which might be a protective mechanism to retain water. The authors also found increased levels of ABA and amino-acid conjugates of IAA, which may contribute to the smaller size of the plants. There’s a LOT of interesting data, so have a look. Your favorite gene or metabolite is likely to appear in this useful and comprehensive study. (Summary by Mary Williams @plantteaching.bsky.social) Plant Cell
Due to anthropomorphic global warming, 2024 was the first year during which the global mean temperature was more than 1.5° above pre-industrial levels. Clearly, understanding how high temperatures affect plant physiology is an urgent priority. In this new study, Zhang et al. did a multi-parameter study of the model C4 plant Setaria viridis. They grew two populations for two weeks at 28°, and then for the next two weeks one group was grown at 42° whilst the other remained at 28°. After this, the authors measured photosynthesis parameters, and carried out metabolomic, transcriptomic, and proteomic studies, leading to several key findings. As expected, the heat-grown plants were significantly smaller than their counterparts. Surprisingly, their photosynthetic efficiency was hardly altered. The heat-grown plants had much higher levels of hexoses, which might be a protective mechanism to retain water. The authors also found increased levels of ABA and amino-acid conjugates of IAA, which may contribute to the smaller size of the plants. There’s a LOT of interesting data, so have a look. Your favorite gene or metabolite is likely to appear in this useful and comprehensive study. (Summary by Mary Williams @plantteaching.bsky.social) Plant Cell