Orange Is the New Green: Arabidopsis ORANGE Represses Chloroplast Biogenesis
Chloroplast development in germinating seedlings initiates upon illumination. Whereas chloroplasts in true leaves develop directly from proplastids, chloroplasts in cotyledons of dark-grown seedlings develop from an intermediate type of plastid called an etioplast. During development, etioplasts accumulate carotenoids and protochlorophyllides in their prolamellar bodies. Once seedlings emerge from the soil into the light, de-etiolation occurs and protochlorophyllides are converted into chlorophylls. Subsequently, the photosystems assemble into thylakoid membranes with the incorporated chlorophylls to form functional chloroplasts (Sundqvist and Dahlin, 1997). Rapid chlorophyll synthesis can result in free chlorophylls that produce highly reactive singlet oxygen, resulting in photodamage. Sun et al., (2019) demonstrate that Arabidopsis thaliana (Arabidopsis) ORANGE (OR), a DnaJ-like zinc finger domain-containing protein, interacts with the bHLH transcription factor TCP14 in the nucleus and regulates both chlorophyll biosynthesis and the development of thylakoid membranes in germinating cotyledons.
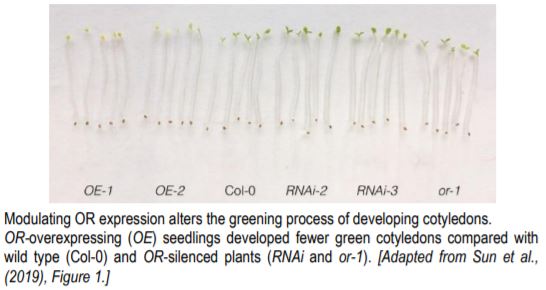
Overexpression of OR in Arabidopsis significantly lowered the percentage of green cotyledons compared with wild type and OR-silenced plants (see figure). Overexpressing lines were delayed in chloroplast biogenesis, and had less chlorophyll, fewer stacks of grana thylakoids, and accumulated more lutein (but less β-carotene). These mutants also had a weaker transcriptional induction and lower abundance of EARLY LIGHT-INDUCIBLE PROTEINS (ELIP1 and ELIP2). The genes encoding these proteins are upregulated when dark-grown seedlings are exposed to light and temporarily bind free chlorophylls (Casazza et al., 2005). In contrast, OR-silenced lines (RNAi silenced or the T-DNA knockout or-1) had a stronger induction of ELIP1 and ELIP2. This indicates that OR negatively regulates chloroplast biogenesis.
 OR is localized to both the chloroplast and nucleus (Sun et al., 2016). Two different antibodies were raised to detect and distinguish between the full-length protein (including chloroplast transit peptide) and the mature OR protein. The authors found that the abundance of full-length OR in the nucleus decreased upon illumination. In the chloroplast, proteins of the inner (TIC) and outer (TOC) chloroplast envelope membranes accumulated 12-24 h post illumination, indicating there was assembly of the TIC/TOC machinery for chloroplast protein import. Full-length OR was found in the plastids of cotyledons up to 12 h post-illumination but was absent in the later stages. By contrast, the mature protein accumulated over time, indicating that removal of the transit peptide from newly imported OR occurs during chloroplast biogenesis. Given the dual localization, the authors targeted OR to the nucleus of the or-1 mutant. The resulting transgenic plants had similar phenotypes as the overexpression mutant lines, indicating that overexpression of the nuclear targeted OR is sufficient to repress de-etiolation.
OR is localized to both the chloroplast and nucleus (Sun et al., 2016). Two different antibodies were raised to detect and distinguish between the full-length protein (including chloroplast transit peptide) and the mature OR protein. The authors found that the abundance of full-length OR in the nucleus decreased upon illumination. In the chloroplast, proteins of the inner (TIC) and outer (TOC) chloroplast envelope membranes accumulated 12-24 h post illumination, indicating there was assembly of the TIC/TOC machinery for chloroplast protein import. Full-length OR was found in the plastids of cotyledons up to 12 h post-illumination but was absent in the later stages. By contrast, the mature protein accumulated over time, indicating that removal of the transit peptide from newly imported OR occurs during chloroplast biogenesis. Given the dual localization, the authors targeted OR to the nucleus of the or-1 mutant. The resulting transgenic plants had similar phenotypes as the overexpression mutant lines, indicating that overexpression of the nuclear targeted OR is sufficient to repress de-etiolation.
By what mechanism does nucleus-localized OR repress chloroplast biogenesis? OR interacts with the bHLH transcription factor TCP14 and represses its transcriptional activity. TCP14 binds to the Up1 element, which is overrepresented in the promoter regions of germination-related genes such as ELIP1 and ELIP2. Using an anti-OR antibody to precipitate chromatin samples, promoter fragments with Up1 were detected in dark-grown wild type seedlings. However, there was no enrichment of Up1-containing fragments of ELIP1 and ELIP2 in tcp14-1 knockout seedlings. These results together suggest that OR affects expression of ELIPs via its interaction with TCP14.
When OR was overexpressed in the tcp14–1 knockout mutant (either by crossing the overexpression mutant with tcp14-1 or transforming tcp14-1 with the OR overexpression construct), seedlings showed greening rates similar to wild type seedlings, and accumulation of ELIPs was rescued. This suggests that the repression of cotyledon greening by OR requires TCP14. The OR-TCP14 interaction in the nucleus represses both ELIP expression and chloroplast biogenesis. When illuminated, the nuclear abundance of OR decreases and consequently, the derepression of TCP14 transcriptional activity results in the etioplast-to-chloroplast transition. This OR-TCP14 interaction helps elucidate the underlying regulatory network required for chloroplast development as plants respond to illumination.
Estee E. Tee
Assistant Features Editor
Australian National University, Canberra
ORCID ID: 0000-0003-2613-5977
REFERENCES
Casazza, AP, Rossini S, Rosso MG and Soave C (2005). Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Mol Biol 58: 41-51.
Sun T, Zhou F, Huang X-Q, Chen W-C, Kong M-J, Zhou C-F, Zhuang Z, Li L and Lu S (2019). ORANGE Represses Chloroplast Biogenesis in Etiolated Arabidopsis Cotyledons via Interaction with TCP14. Plant Cell. Published Dec. 2019. DOI: https://doi.org/10.1105/tpc.18.00290.
Sun T, Zhou F, Liu C-J, Zhuang Z and Lu S (2016). The DnaJ-like zinc finger domain protein ORANGE localizes to the nucleus in etiolated cotyledons of Arabidopsis thaliana. Protoplasma 253: 1599-1604.
Sundqvist C and Dahlin C (1997). With chlorophyll pigments from prolamellar bodies to light-harvesting complexes. Physiol Plant 100: 748-759.