New insights into Marchantia polymorpha genome
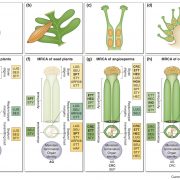
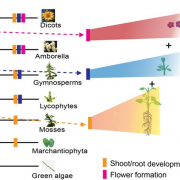
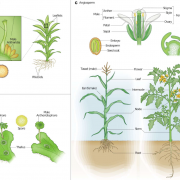
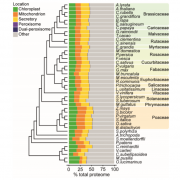
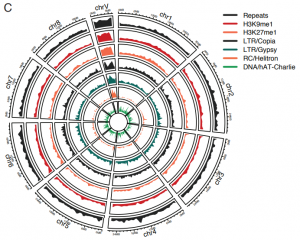 Since the publication of the Marchantia polymorpha genome in 2017, a big update was necessary. In the past week, two groups published on improving Marchantia chromosome assembly following different experimental approaches. Diop et al. made a high-density linkage map using genetic markers obtained by ddRAD-seq, managing to assemble 88% of the genomic DNA content in near-complete chromosomes. On the other hand, Montgomery et al. additionally made Hi-C analysis and achieved to map 98.6% of genomic DNA in chromosomes. Both approaches arrived at similar conclusions about the relative localization of transposable elements (TE) and other gene features. While in angiosperms centromeres are enriched in TE and depleted in protein-coding genes, in Marchantia most gene features are evenly distributed across the chromosome. This also was demonstrated for the model moss Physcomitrella patens, suggesting that this is a conserved feature of bryophytes. Additionally, Montgomery et al. performed CUT&RUN, drawing many interesting conclusions particularly related to H3K27me3 in the spatial and linear organization of the Marchantia genome. Diop et al. instead analyzed the distribution of recombination rates based on crosses with a Switzerland accession. In conclusion, both papers offer new insights into how chromosome architecture has evolved in eukaryotes. Additionally, they provide useful genomic resources to the community working with Marchantia. (Summary by Facundo Romani) (10.1101/827881 & 10.1111/TPJ.14602)
Since the publication of the Marchantia polymorpha genome in 2017, a big update was necessary. In the past week, two groups published on improving Marchantia chromosome assembly following different experimental approaches. Diop et al. made a high-density linkage map using genetic markers obtained by ddRAD-seq, managing to assemble 88% of the genomic DNA content in near-complete chromosomes. On the other hand, Montgomery et al. additionally made Hi-C analysis and achieved to map 98.6% of genomic DNA in chromosomes. Both approaches arrived at similar conclusions about the relative localization of transposable elements (TE) and other gene features. While in angiosperms centromeres are enriched in TE and depleted in protein-coding genes, in Marchantia most gene features are evenly distributed across the chromosome. This also was demonstrated for the model moss Physcomitrella patens, suggesting that this is a conserved feature of bryophytes. Additionally, Montgomery et al. performed CUT&RUN, drawing many interesting conclusions particularly related to H3K27me3 in the spatial and linear organization of the Marchantia genome. Diop et al. instead analyzed the distribution of recombination rates based on crosses with a Switzerland accession. In conclusion, both papers offer new insights into how chromosome architecture has evolved in eukaryotes. Additionally, they provide useful genomic resources to the community working with Marchantia. (Summary by Facundo Romani) (10.1101/827881 & 10.1111/TPJ.14602)