Molecular teamwork: how three protein families and DNA work together to control gene expression
Hong and Rusnak et al. explore how a 1000-bp region of DNA activates gene expression in giant cells.
By Adrienne Roeder and Byron Rusnak
Background: Specialized cell types carry out specific functions within plants. Some genes are expressed uniquely in one cell type and not expressed in other cells; these genes may be important for the function of that cell type. These cell type-specific expression patterns are created by transcription factors that bind to regions of the DNA called enhancers to regulate when and where a gene is expressed. Yet the regulatory logic behind how transcription factors that are more broadly expressed combine to create a cell type-specific expression pattern is not well understood. For example, giant cells are highly enlarged cells in the Arabidopsis sepal that control sepal curvature and promote defense from pathogens and insects. We previously identified a 1000-bp region of DNA that is sufficient to turn on the expression of a reporter gene specifically in giant cells.
Question: We asked how this 1000-bp DNA region activates gene expression specifically in giant cells.
Findings: Using Arabidopsis, we found that the combined activity of transcription factors from three families are important for generating the giant cell-specific pattern. TCP-type transcription factors promote expression from the enhancer in all epidermal cells. The transcription factor ATML1 modulates the expression level driven by the enhancer. As ATML1 protein is highly accumulated in giant cells, it can bind to the enhancer and drive high expression in giant cells. Dof transcription factors repress expression by binding to the enhancer, resulting in high expression in giant cells only. We created a conceptual model for how transcription factors whose encoding genes are expressed broadly in many cell types can bind to the same enhancer, combining their activities to produce cell type-specific expression patterns.
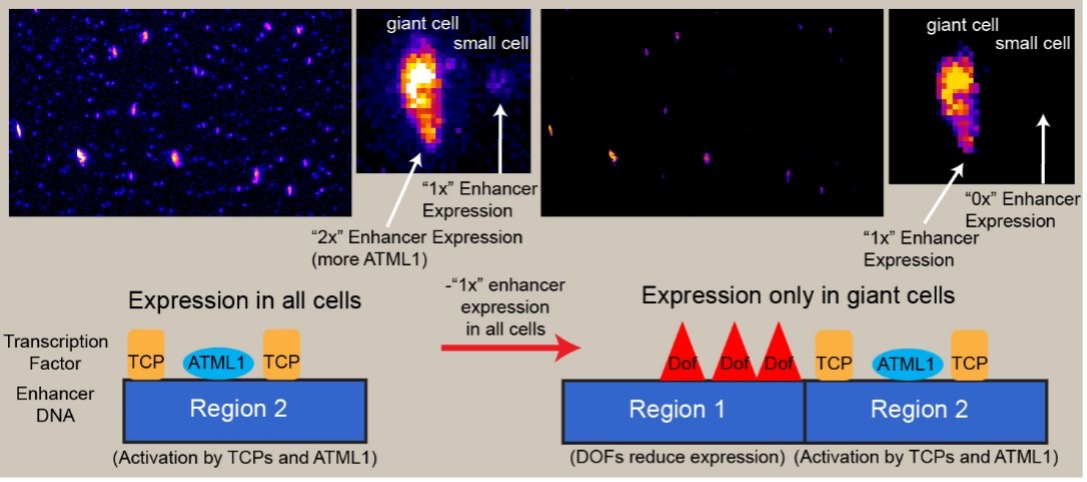
Model for how three different kinds of transcription factors combine on the enhancer to create giant cell specific expression.
Next steps: A future challenge will be to use this information as a basis for engineering new synthetic enhancers with cell type-specific expression.
Reference:
Lilan Hong, Byron Rusnak, Clint S. Ko, Shouling Xu, Xi He, Dengying Qiu, S. Earl Kang, Jose L. Pruneda-Paz, Adrienne H. K. Roeder. (2023). Enhancer activation via TCP and HD-ZIP and repression by Dof transcription factors mediate giant cell-specific expression. https://doi.org/10.1093/plcell/koad054