Methionine-Derived Glucosinolates: The Compounds that Give Brassicas their Bite
The compounds that give wasabi (Eutrema japonicum) its kick and bok choy (Brassica rapa ssp. chinensis) its distinctive flavor are breakdown products of glucosinolates derived from not so pungent amino acids. These zesty phytochemicals help Brassica plants adapt to their many environmental niches worldwide (reviewed in Halkier and Gershenzon, 2006). While these compounds are craved by many and serve as crop protectants, high glucosinolate contents in oilseed-derived animal feed can be toxic, which prompted breeders to reduce their levels. The diversity of the almost 130 glucosinolates in Brassicas, their scores of breakdown products, and the many associated enzymes has made studying their biosynthesis and evolution a daunting task.
The biosynthesis of the highly abundant aliphatic methionine-derived glucosinolates involves three steps: side-chain elongation, core structure formation, and side-chain modification. During side-chain elongation, acetyl-coenzyme-A (acetyl-CoA) is condensed to a methionine-derived 2-oxo acid substrate to form a substituted 2-malate derivative. This committed step of the pathway is catalyzed by methylthioalkylmalate synthase (MAMS) (Kliebenstein et al., 2001). The number of times MAMS catalyzes elongation determines the length of the aliphatic side-chain, but how this gatekeeper function of MAMS evolved in Brassica crops is poorly understood due to the polyploid nature of these plants and the neo-functionalization of paralogous genes during evolution.
 Kumar et al. (2019) probed the genetics, biochemistry, and structural biology of MAMS proteins in Brassicas to unravel the molecular basis for their evolution and the diversification of methionine-derived glucosinolates. The study began with phylogenetic analysis of MAM-like genes from several diploid Brassica species identified based on sequence homology with genes from Arabidopsis thaliana. Brassica genomes encode multiple MAMS proteins with different gene expression profiles and preferences for 2-oxo substrates, accounting for the diversity of aliphatic glucosinolates across Brassicas. The major oilseed crop Brassica juncea, an allotetraploid species containing the genomes of the mustards B. rapa and B. nigra, harbors four MAMS genes that fall into two subclades with different expression profiles: Bj-MAM1 and Bj-MAM2. The transcript levels of Bj-MAMS genes derived from B. rapa tend to be higher than those derived from B. nigra, pointing to transcriptional dominance in B. juncea.
Kumar et al. (2019) probed the genetics, biochemistry, and structural biology of MAMS proteins in Brassicas to unravel the molecular basis for their evolution and the diversification of methionine-derived glucosinolates. The study began with phylogenetic analysis of MAM-like genes from several diploid Brassica species identified based on sequence homology with genes from Arabidopsis thaliana. Brassica genomes encode multiple MAMS proteins with different gene expression profiles and preferences for 2-oxo substrates, accounting for the diversity of aliphatic glucosinolates across Brassicas. The major oilseed crop Brassica juncea, an allotetraploid species containing the genomes of the mustards B. rapa and B. nigra, harbors four MAMS genes that fall into two subclades with different expression profiles: Bj-MAM1 and Bj-MAM2. The transcript levels of Bj-MAMS genes derived from B. rapa tend to be higher than those derived from B. nigra, pointing to transcriptional dominance in B. juncea.
Purified Bj-MAMS proteins expressed in Escherichia coli were incubated with acetyl-CoA and 2-oxo substrates and their products monitored by liquid chromatography–mass spectrometry. These proteins showed various levels of activity. Kinetic analysis of Bj-MAMS incubated with acetyl-CoA and various glucosinolate precursors revealed that the two Bj-MAMS subclades have different substrate preferences.
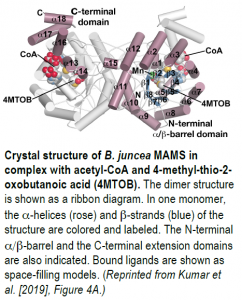
To further probe MAMS activity, the authors crystallized BjMAM1-A in complex with Mn2+, acetyl-CoA, and the 2-oxo substrate, 4-methyl-thio-2-oxobutanoic acid, and solved its x-ray crystal structure at 2.1 Å resolution (see figure). BjMAM1-A is a dimeric protein belonging to the DRE-TIM metallolyase enzyme superfamily with a binding pocket for 4-methyl-thio-2-oxobutanoic acid and Mn2+ deep in its fold. The two sets of Bj-MAMS isoforms differ by just four amino acids in this region, which dictate specificity for different sized 2-oxo substrates, as confirmed by biochemical analysis of mutated MAMS proteins. Pinpointing these crucial amino acids sheds light on glucosinolate diversity and paves the way for altering the contents of glucosinolates and their pungent metabolites in Brassica crops, which would benefit plants and humans alike.
REFERENCES
Halkier, B.A. and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Ann. Rev. Plant Biol. 57: 303-333.
Kliebenstein, D., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., and Mitchell-Olds, T. (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126: 811-825.
Kumar, R., Lee, S.G., Augustine, R., Reichelt, M., Vassão, D., Palavalli, M.H., Allen, A., Gershenzon, J., Jez, J.M., and Bisht, N.C. (2019). Molecular basis of the evolution of methylthioalkylmalate synthase and the diversity of methionine-derived glucosinolates. Plant Cell 31: doi:10.1105/tpc.19.00046. (https://doi.org/10.1105/tpc.19.00046)