Meiocyte-specific Small RNAs and Meiotic Recombination: Questions and Anthers
Small RNAs regulate gene expression and epigenetic modifications (via RNA-directed DNA methylation), and therefore play key roles in plant development (reviewed in Borges and Martienssen, 2015). Moreover, emerging evidence indicates that small RNAs may have a role in the repair of double-stranded breaks (DSB) induced by DNA damage or nuclease cleavage, as 21-nucleotide small RNAs are produced from sites near DSBs (reviewed in Hawley et al., 2017). Repair of DSBs has major importance for genome stability and is key for meiosis, when the cell generates DSBs during recombination. Many studies have examined the relationship between small RNAs and artificially induced DSBs, but the link between developmentally programmed DSBs and small RNAs remains an outstanding question.
Nevertheless, to quote the cartoon science teacher Ms. Frizzle, “For every question, there is an anther.” If your question was “What is the link between small RNAs, DSBs, and meiotic recombination?” then new work in The Plant Cell has your answer, and anther. Finding answers in anthers is challenging because only a few cells in plant anthers undergo meiosis; however new studies have addressed epigenetic regulation in meiosis (reviewed in Martinez and Köhler, 2017) by coupling novel methods to identify and isolate specific cell types with high-throughput sequencing technologies that can yield good data from very few cells. In the featured paper, Huang et al. (2019) use a capillary method, where they scoop up meiocytes from squashed anthers, to purify enough Arabidopsis thaliana meiocytes for high-throughput sequencing of mRNAs and small RNAs.
Using this method, the authors identify a set of 23- and 24-nucleotide small RNAs and mapping these small RNAs to the Arabidopsis thaliana genome reveals clusters at thousands of sites in the genome. Moreover, by comparing leaf and meiocyte small RNA clusters, the authors define about two thousand meiocyte-specific small RNA (ms-sRNA) clusters. These ms-sRNAs require RNA polymerase IV and Dicer function, but the authors note that until ms-sRNA biogenesis is elucidated, they should not be further classified as microRNAs or small interfering RNAs. In somatic cells, small RNA clusters mostly map to heterochromatin and transposable elements; by contrast, the ms-sRNA clusters mostly map to coding sequences. Moreover, comparison of the ms-sRNAs and the mRNA sequencing data showed that ms-sRNAs tended to associate with the gene bodies of expressed genes, in contrast to somatic cells where small RNAs tend to associate with silenced genes. Indeed, the authors identified 149 ms-sRNA-associated genes whose expression was specifically upregulated in meiocytes, including two genes encoding products involved in recombination, the recombinase RAD51 and the E3 ubiquitin ligase component ARABIDOPSIS SKP1-LIKE1. The observed changes in gene expression were not related to differential methylation between leaf cells and meiocytes.
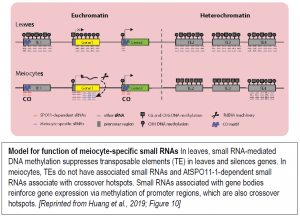 Meiocytes undergo a complex, dynamic developmental process that likely involves many different small RNA functions. To examine the connection between ms-sRNAs and meiotic recombination, the authors examined the small RNA profiles of mutants defective in DSB formation due to lack of SPORULATION 11-1 (SPO11-1). The spo11-1-1 meiocytes lacked about two-thirds of the small RNA clusters found in wild-type meiocytes. Intriguingly, the SPO11-1-dependent ms-sRNA clusters tended to associate with crossover hotspot-associated motifs (CTT repeats and A-rich motifs) and with hotspot-associated defense-related genes, particularly those encoding nucleotide-binding sequence-leucine-rich repeat (NBS-LRR) proteins with a CNN motif. Based on these observations, the authors propose a model for the functions of the ms-sRNAs (figure).
Meiocytes undergo a complex, dynamic developmental process that likely involves many different small RNA functions. To examine the connection between ms-sRNAs and meiotic recombination, the authors examined the small RNA profiles of mutants defective in DSB formation due to lack of SPORULATION 11-1 (SPO11-1). The spo11-1-1 meiocytes lacked about two-thirds of the small RNA clusters found in wild-type meiocytes. Intriguingly, the SPO11-1-dependent ms-sRNA clusters tended to associate with crossover hotspot-associated motifs (CTT repeats and A-rich motifs) and with hotspot-associated defense-related genes, particularly those encoding nucleotide-binding sequence-leucine-rich repeat (NBS-LRR) proteins with a CNN motif. Based on these observations, the authors propose a model for the functions of the ms-sRNAs (figure).
This study adds a new dimension to the role of small RNAs in DSB repair, but intriguing questions (many raised in the Hawley et al., 2017 review) remain open for further work, including the mechanistic role of these small RNAs and how they are produced– perhaps by degradation of a longer RNA that functions in DSB repair. Moreover, some future questions may take us beyond anthers by examining the role of these small RNAs in female meiocytes, an even trickier cell type to examine. With all these questions, it is to be hoped that these authors continue working on the anthers.
REFERENCES
Borges, F., and Martienssen, R.A. (2015). The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16, 727-741.
Ben R Hawley, Wei-Ting Lu, Ania Wilczynska, and Martin Bushell (2017) The emerging role of RNAs in DNA damage repair. Cell Death Differ. 2017 Apr; 24(4): 580–587. doi: 10.1038/cdd.2017.16
Jiyue Huang, Cong Wang, Haifeng Wang, Pingli Lu, Binglian Zheng, Hong Ma, Gregory P. Copenhaver, and Yingxiang Wang (2019). Meiocyte-specific and AtSPO11-1-dependent Small RNAs and Their Association with Meiotic Gene Expression and Recombination. Plant Cell DOI: https://doi.org/10.1105/tpc.18.00511.
German Martinez and Claudia Köhler (2017) Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Current Opinion in Plant Biology Volume 36, April 2017, Pages 22-28. https://doi.org/10.1016/j.pbi.2016.12.006



