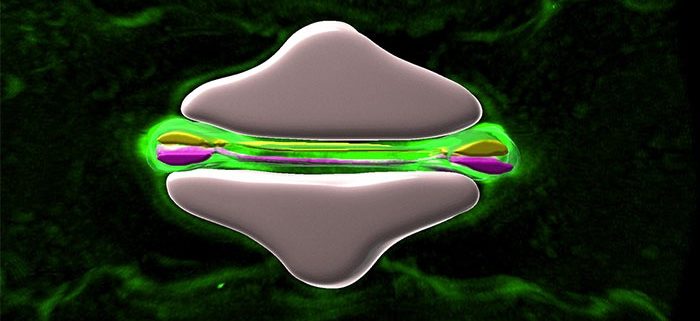
Maize stomata during movement and development
Sun et al. explore the molecular basis of the stomatal response and stomatal development in maize. https://doi.org/10.1093/plcell/koac047
Guiling Sun, State Key Laboratory of Crop Stress Adaptation and Improvement, State Key Laboratory of Cotton Biology, School of Life Sciences, Henan University
David Galbraith, School of Plant Sciences and Bio5 Institute, The University of Arizona
Chun-Peng Song, State Key Laboratory of Crop Stress Adaptation and Improvement, State Key Laboratory of Cotton Biology, School of Life Sciences, Henan University
Background: As the outmost layer of cells, the leaf epidermis controls gas and water exchange between the plant and its environment, and provides protection from various biotic and abiotic stresses. Within the epidermis, stomata determine photosynthetic and water-use efficiencies, responding rapidly to changing environmental conditions. Unlike dicot stomata, grass stomata contain two subsidiary cells (SCs) flanking two dumbbell-shaped guard cells (GCs). Coordinated regulation of SCs and GCs allows higher speeds of stomatal movement in response to environmental changes, and results in higher water-use efficiencies.
Question: Can we obtain a comprehensive understanding concerning the molecular basis underlying the fast stomatal response and stomatal development in grass? What are the key players in stomata movement and development, such as membrane channels, components of key signaling pathways, and GC or SC cell wall-specific proteins?
Findings: We firstly isolated propidium iodide-stained nuclei from maize epidermal peels by fluorescence-activated sorting, and developed a high-throughput platform of single nucleus RNA sequencing. Assisted by reporter lines involving transgenic maize and rice, we acquired the transcriptional profiles of the different cell types in maize peels, including GCs, SCs, epidermal cells, the attached mesophyll, and vascular-related cells. We found GC- and SC-specific genes encoding previously reported and new transporters, components of abscisic acid, CO2, Ca2+, starch metabolism, and of blue light signaling pathways identified in other species. Using the same technical approach, we analyzed samples of the maize leaf base sample, obtaining new candidates involved in grass stomatal development. Among these candidates, we identified cell wall-related genes that may play important roles in dumbbell-shaped morphogenesis of developing and mature stomata.
Next steps: Functional experiments are needed to test the hypotheses emerging from the single nucleus transcriptome data. Transcriptomic comparison of maize stomata with other monocot crops would provide deeper insights into the evolutionary innovation of the dumbbell-shaped stomata.
Guiling Sun, Mingzhang Xia, Jieping Li, Wen Ma, Qingzeng Li, Jinjin Xie, Shenglong Bai, Shanshan Fang, Ting Sun, Xinlei Feng, Guanghui Guo, Yanli Niu, Jingyi Hou, Wenling Ye, Jianchao Ma, Siyi Guo, Hongliang Wang, Yu Long, Xuebin Zhang, Junli Zhang, Hui Zhou, Baozhu Li, Jiong Liu, Changsong Zou, Hai Wang, Jinling Huang, David W. Galbraith, Chun-Peng Song. (2022). The maize single-nucleus transcriptome comprehensively describes signaling networks governing movement and development of grass stomata. https://doi.org/10.1093/plcell/koac047