Linking low energy sensing to chromatin modification and gene activity in rice
Wang and colleagues investigated the starvation response of rice seedlings.
By Wentao Wang and Yue Lu
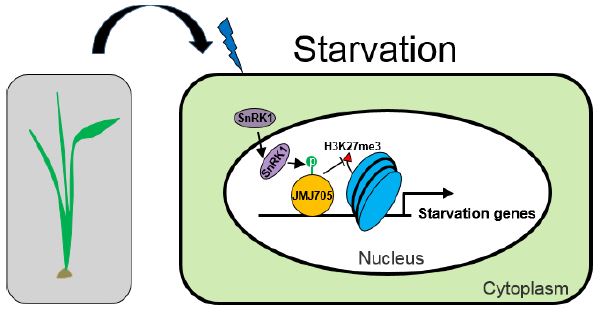
Background: For survival and growth, cells of all living organisms must balance their energy consumption and supply. Although energetically autonomous (through photosynthesis), plants are continuously challenged by their changing environment and especially by diverse stresses that affect plant physiology and cellular energy supply. The evolutionarily conserved protein complex, called SnRK1 in plants, senses cellular energy levels. SnRK1 is a protein kinase that phosphorylates other proteins to regulate their activity. When cellular energy levels become low, SnRK1 can activate energy producing- and represses energy consuming-reactions by regulating the expression of many genes. Gene expression (activity) depends on the presence of specific transcription factors and the structure of chromatin (assembly of DNA and nuclear proteins such as histones) at promoter regions.
Question: Being a protein kinase, how can SnRK1 activate or repress many genes in cells under starvation? Does SnRK1 alter chromatin structure of energy-response genes, and if so how does SnRK1 behave when cellular energy supply is sufficient?
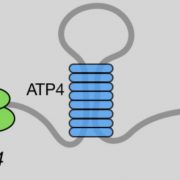
Findings: In rice seedlings under starvation conditions, SnRK1 is enriched in the nucleus, where it interacts with and phosphorylates the histone demethylase (which removes methyl groups from histone proteins) called JMJ705. Covalent modification of histone H3 on lysine 27 with three methyl groups (H3K27me3) represses the transcription of genes that associate with histones bearing this mark. The phosphorylation by SnRK1 stimulates JMJ705 activity to remove H3K27me3 from chromatin (or histones) of key starvation-responsive genes, among which are those encoding known starvation-responsive transcription factors. Thus, the SnRK1-JMJ705 pathway identified in this work links cellular energy sensing to chromatin modification and gene expression. In addition, we find that in cellular energy-sufficient conditions, SnRK1 and JMJ705 are required to repress the starvation-induced gene expression program.
Next steps: Future research will focus on the precise mechanisms by which SnRK1 and JMJ705 activate the starvation-induced gene expression program under stress and repress the program in an energy-sufficient state. This research may contribute to improving crop yield by reducing the trade-off between stress tolerance and growth.
Wentao Wang, Yue Lu, Junjie Li, Xinran Zhang, Fangfang Hu, Yu Zhao, Dao-Xiu Zhou. (2021). SnRK1 stimulates the histone H3K27me3 demethylase JMJ705 to regulate a transcriptional switch to control energy homeostasis. https://doi.org/10.1093/plcell/koab224