Key factors that interactively regulate maize endosperm development
Wu et al. explore how three key transcription factors co-regulate gene networks and the regulatory landscape associated with endosperm development and seed phenotype in maize.
https://doi.org/10.1093/plcell/koad247
Hao Wu1,2 and Philip W. Becraft1,3
- Iowa State University, Genetics, Development & Cell Biology Department, Ames, IA;
- Cornell University (Current), School of Integrative Plant Science, Ithaca, NY;
- Iowa State University, Agronomy Department, Ames, IA
Background: The endosperm of cereal plants such as rice (Oryza sativa) and maize (Zea mays) serves as a storage tissue for nutrients, supplying energy for germination and the initial growth of seedlings. It is also a source for human food, livestock feed, and industrial applications. The endosperm develops in two stages, a cellular development phase characterized by extensive cell division and cell differentiation, and a second phase, grain filling, in which these cells accumulate storage materials, particularly starches and proteins. Three key transcription factors, NAKED ENDOSPERM1 (NKD1), NKD2, and OPAQUE2 (O2), play important roles in maize endosperm cellular development and storage material accumulation. The mutants of these factors showed several common defects in endosperm grain-filling, and this suggests that they may have some related functions. However, we know little about how the three factors interactively regulate downstream genes and the regulatory landscapes that modulate endosperm development.
Question: How do NKD1, NKD2 and O2 co-regulate gene networks and the regulatory landscape associated with endosperm development and how do they interact to control seed phenotype?
Findings: We developed a set of homozygous lines consisting of nkd1, nkd2 and o2 mutants in all single, double, and triple mutant combinations, as well as wild type. We identified synthetic phenotypes suggesting that the three factors interactively affect endosperm development. We found that the three factors may constrain hormone responses, cell wall organization, as well as other cellular developmental processes, and promote starch metabolism, lipid storage, and storage protein accumulation during the transition from cellular development to storage compound accumulation. These processes are regulated through a dynamic, hierarchical gene network and the three factors function as the central regulators. We also reported potential direct target genes of NKD1 and NKD2 along with their chromatin accessibility status and differentially expressed genes between wild type and each mutant.
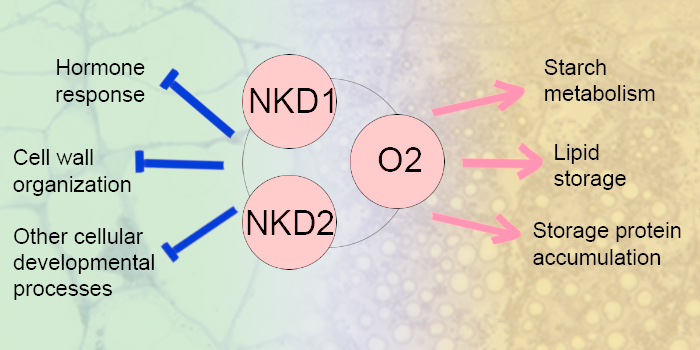
Next steps: In the future, we plan to test some key direct targets of NKD1 and NKD2 by developing mutant lines to further explore how they work together to regulate endosperm development.
Reference:
Hao Wu, Mary Galli, Carla J. Spears, Junpeng Zhan, Peng Liu, Ramin Yadegari, Joanne M. Dannenhoffer, Andrea Gallavotti, Philip W. Becraft (2023). NAKED ENDOSPERM1, NAKED ENDOSPERM2, and OPAQUE2 interact to regulate gene networks in maize endosperm development. https://doi.org/10.1093/plcell/koad247