Integrating Coexpression Networks with GWAS
Schaefer et al. integrate co-expression networks from three diverse maize gene expression data sets with genome wide association study data to prioritize genes related to the maize grain ionome. Plant Cell https://doi.org/10.1105/tpc.18.00299
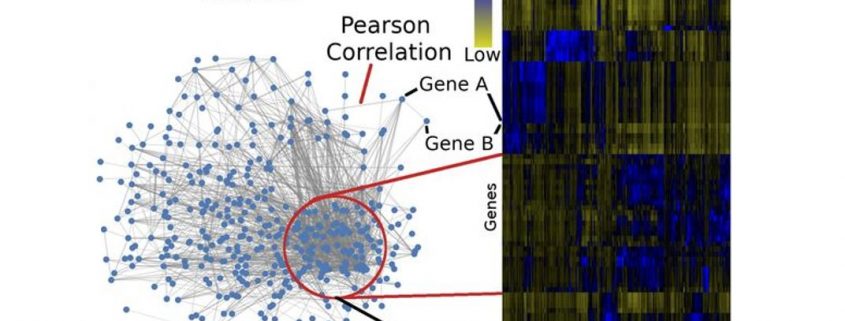
Background: Genetics examines the relationships between DNA and the physical traits of organisms. While we can accurately describe simple traits with a small number of genes, complex traits are harder to explain and are thought to be controlled by tens or hundreds of genes. In many agricultural species, this problem is even more pronounced, as the functions of many genes are unknown. To help mitigate this issue, we organized data in the form of a biological network, creating profiles for each gene using data from different sources. Like social networks, where you might predict what kind of movies a person likes based on their music tastes, we predict which traits a gene might be important for by looking at which tissues they are in and under what conditions they are active.
Question: In this study, we wanted to test what data was best to create biological networks and to build software to do the analysis. We analyzed results from a genetics experiment that looked at the relationship between DNA and the nutritional components of Zea mays, commonly known as maize or field corn.
Findings: Alongside the software tools to build networks, we also created tools to measure the “health” of the networks before we used them to analyze and genetic data. Using simulated genetic studies, we tested that predictions made about gene functions were accurate and meaningful. We found that networks constructed from different tissues or maize varieties had a substantial impact in their utility to analyze genetic data. Using our method, we analyzed the results of an experiment looking at the genetics of maize nutritional quality. By combining the networks with the genetic data, we filtered our candidate list of genes from tens of thousands of genes to 610 high priority genes. Furthermore, we show that our method and software are generalizable and can be used to analyze other genetic datasets.
Next steps: In the future, we will apply this method to other interesting traits in maize and other plant species, as well as applying the approach to help analyze genetics experiments in animal species.
Robert J. Schaefer, Jean-Michel Michno, Joseph Jeffers, Owen Hoekenga, Brian Dilkes, Ivan Baxter, Chad L. Myers. (2018). Integrating Coexpression Networks with GWAS to Prioritize Causal Genes in Maize. Plant Cell 30: 2922-2942; DOI: https://doi.org/10.1105/tpc.18.00299