In the Histone Zone: The Mighty Eraser
Histones undergo myriad covalent modifications – more than 100 types have been identified in the 50+ years since Allfrey, Faulkner, and Mirsky (1964) found that increased histone acetylation was associated with genomic regions of active transcription (reviewed in Zentner and Henikoff, 2013). Enzymes referred to as “writers” add chemical modifications on DNA or histones and “erasers” remove these modifications. Different types of histone modifications tend to be found at different cis-regulatory elements (such as promoters and enhancers), but the mechanisms regulating their presence remain unclear.
One key modification, histone methylation, involves the addition of one, two, or three methyl groups to a lysine or arginine and affects transcription. For example, methylation of lysines 4 or 36 on histone H3 (H3K4me3 and H3K36me2 or 3) is associated with increased transcription, and H3K9me3 (or H3K9me2 in plants) and H3K27me3 are associated with repressed transcription, including formation of heterochromatin. Histone demethylases erase the methylation marks and the specificity of these enzymes thus has major implications for gene regulation. Yang et al. (2018) provide insight on this specificity by decoding the crystal structure of the Arabidopsis thaliana Lysine (K)-Specific Demethylase 5 (KDM5) subfamily H3K4me3 demethylase JUMONJI 14 (JMJ14). JMJ14 has essential functions in the regulation of flowering time, RNA-directed DNA methylation, and gene expression; in humans, enzymes in the KDM5 subfamily have potential as targets for anti-cancer drugs.
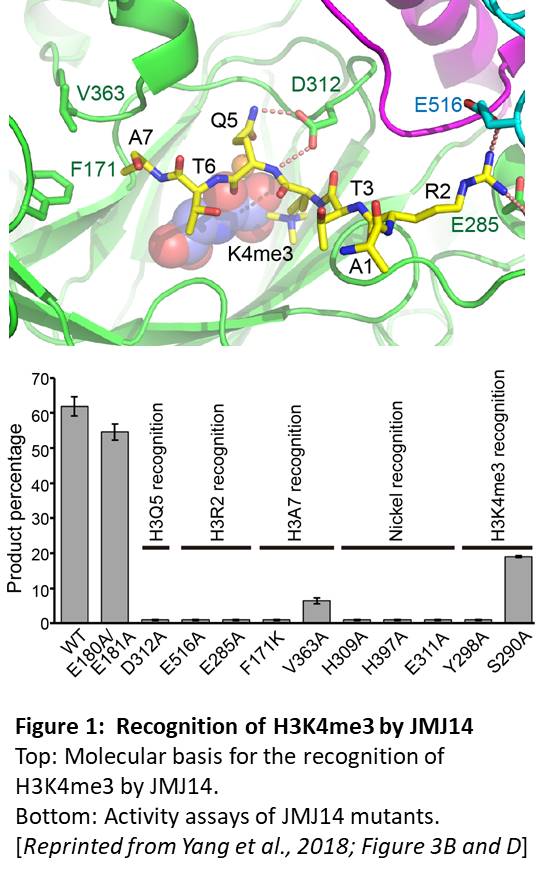 The authors first used a mass spectrometry-based assay for demethylation to show that a truncated version of JMJ14 specifically demethylated H3K4me3, but not H3K9me2, H3K27me3, or H3K36me3 marks. The crystal structures of JMJ14 (free or bound to a 10-amino acid peptide from H3, with H3K4me3 modification), showed that recognition of the R2 and Q5 residues of H3 by the E285, E516, and D312 residues of JMJ14 contributes to substrate specificity (Figure 1, top). Moreover, mutants that change the key JMJ14 residues to alanine have significantly decreased the demethylase activity (Figure 1, bottom) in vitro and in Nicotiana benthamiana leaves. Intriguingly, these acidic residues are conserved in the corresponding human demethylase and mutation of these residues causes loss or elimination of activity of the human enzyme, indicating that study of the plant enzymes may have implications for understanding (and possibly targeted drug design towards) the human enzymes.
The authors first used a mass spectrometry-based assay for demethylation to show that a truncated version of JMJ14 specifically demethylated H3K4me3, but not H3K9me2, H3K27me3, or H3K36me3 marks. The crystal structures of JMJ14 (free or bound to a 10-amino acid peptide from H3, with H3K4me3 modification), showed that recognition of the R2 and Q5 residues of H3 by the E285, E516, and D312 residues of JMJ14 contributes to substrate specificity (Figure 1, top). Moreover, mutants that change the key JMJ14 residues to alanine have significantly decreased the demethylase activity (Figure 1, bottom) in vitro and in Nicotiana benthamiana leaves. Intriguingly, these acidic residues are conserved in the corresponding human demethylase and mutation of these residues causes loss or elimination of activity of the human enzyme, indicating that study of the plant enzymes may have implications for understanding (and possibly targeted drug design towards) the human enzymes.
In contrast to methylation, which does not alter the positive charge of lysine, acetylation of specific lysines on histones neutralizes the positively charged lysine. Histone deacetylation changes the accessibility of chromatin and has roles in processes such as DNA replication, transcription, and possibly DNA repair (reviewed in Zentner and Henikoff, 2013). Chen et al. (2018) examined the closely related Arabidopsis thaliana histone deacetylases HD2B and HD2C, which interact to form an oligomeric complex in vivo. Overexpression of HD2C reduced the global acetlyation levels (H4K16ac, and to a lesser extent H4K8ac, and H3K23ac, but not H3K9ac) in Nicotiana benthamiana and depletion of HD2C induced hyperacetylation of H4K16 in Arabidopsis thaliana. Therefore, these factors affect histone acetylation genome-wide.
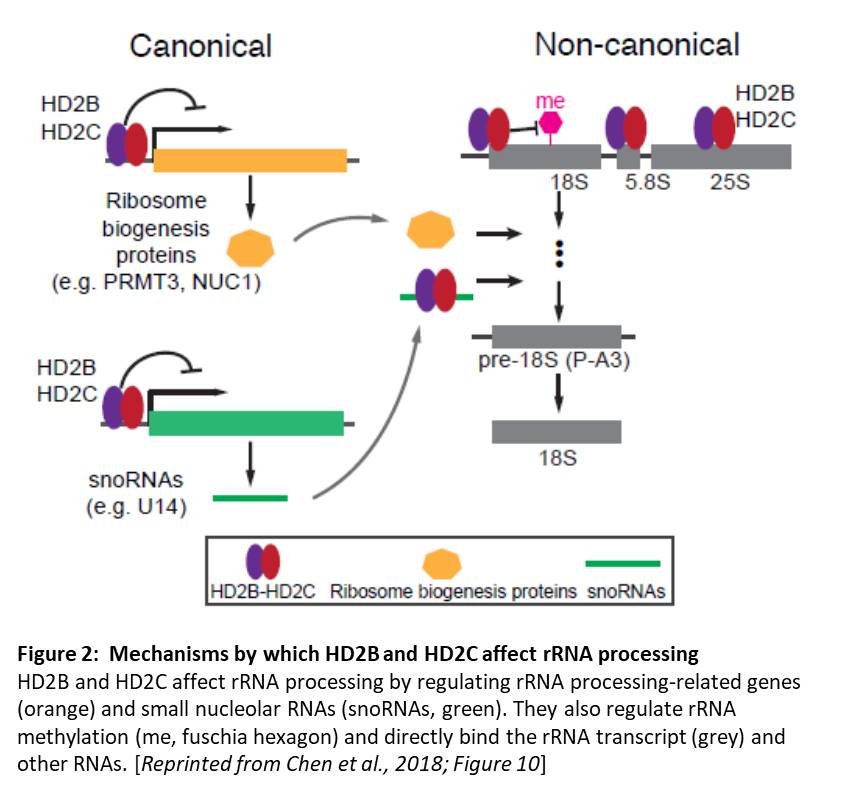
Identification of the specific genes targeted by these deacetylases showed a previously unidentified link to ribosomal RNA processing (rRNA). A genome-wide scan of HD2C binding sites showed that HD2C is enriched in the promoters of active genes. The set of genes bound by HD2C and affected by the changes in acetylation in HDC2 depletion lines was enriched in genes related to ribosome biogenesis. Moreover, hd2b hd2c double mutants showed the hallmarks of ribosome deficiencies and accumulated unprocessed 18S rRNA. The double mutants showed normal levels of rRNA transcripts, indicating no effect on rDNA silencing, but did show perturbed levels of transcripts related to rRNA processing and increased levels of methylation of the rRNAs. HD2B and HD2C localize to the nucleolus and directly bind rRNA transcripts, indicating that their effect on rRNA processing is not just an indirect effect via gene regulation. Intriguingly, HD2B and HD2C also associate with other RNAs, including mRNAs and small nucleolar RNAs. Therefore, this study identified canonical (via regulation of rRNA processing-related genes) and non-canonical (effects on rRNA methylation and binding to the rRNA transcript) mechanisms by which HD2B and HD2C affect rRNA processing (Figure 2).
In sum, the pen may be mightier than the sword, but these studies show the importance of the eraser.
REFERENCES
Allfrey, C.L., Faulkner, R., and Mirsky, A.E. (1964) Acetylation and methylations of histones and their possible roles in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA. 51 786–794.
Chen, X., Lu, L., Qian, S., Scalf, M., Smith, L.M., Zhong, X. (2018) Canonical and Non-canonical Actions of Arabidopsis Histone Deacetylases in Ribosomal RNA Processing. Plant Cell. DOI: 10.1105/tpc.17.00626.
Yang Z., Qiu, Q., Chen, W., Jia, B., Chen, X., Hu, H., He, K., Deng, X., Li, S., Tao, W.A., Cao, X., Du, J. (2018). Structure of the Arabidopsis JMJ14–H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases. Plant Cell. DOI: 10.1105/tpc.17.00666.
Zentner, G.E. and Henikoff, S. (2013) Regulation of nucleosome dynamics by histone modifications Nature Structural & Molecular Biology 20, 259–266. doi:10.1038/nsmb.2470