Inhibition of TOR, Nitrogen Assimilation, and Amino Acid Biosynthesis: Lessons from Chlamydomonas
To survive, organisms must sense their nutritional status (including nutrient availability and quality) and regulate their growth and metabolism accordingly. In plants, animals, and fungi, the Target of Rapamycin (TOR) kinase regulates metabolism, nutrient sensing, and growth (reviewed in Dobrenel et al., 2016). However, in most plants, rapamycin does not efficiently inhibit TOR function, hampering studies of its upstream and downstream factors; by contrast, rapamycin does inhibit Chlamydomonas reinhardtii TOR and therefore, studies of green algae have much to tell us about TOR function in photosynthetic organisms.
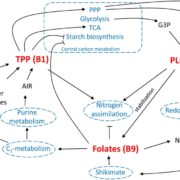
One key balance that TOR regulates is the switch between catabolic metabolism (such as the induction of autophagy), and anabolic metabolism (such as the induction of biosynthetic pathways). Indeed, previous work showed that treatment with rapamycin induces accumulation of starch and triacylglycerols, and a surprisingly rapid accumulation of free amino acids. Where do all these amino acids come from? Potential explanations included the inhibition of translation, or induction of autophagy. To examine this question, Mubeen et al. (2018) conducted a targeted metabolic profiling of Chlamydomonas cells grown under photoautotrophic conditions. In these cells, treatment with rapamycin or the TOR inhibitor AZD-8055 induced substantial accumulation of almost all amino acids in the first five minutes of TOR inhibition. Moreover, this increase still occurred in cells treated with the translational inhibitor cycloheximide or the proteasome inhibitor MG-132, indicating that the previously suggested explanations (inhibition of translation or induction of proteolysis) only partially contribute to this accumulation. By contrast, restriction of fixed carbon (in dark-incubated cells) or nitrogen (in the media) limited the accumulation of amino acids following rapamycin treatment, although a subset of amino acids did still accumulate in nitrogen-limited cells.
The results of the nutrient limitation experiments indicated that nitrogen uptake or assimilation might be affected by rapamycin. To test this, the authors used isotope labeling with 15NH4+ to examine N uptake and assimilation. Indeed, compared with untreated controls, the rapamycin-treated cells showed higher cellular levels of 15N accumulation in amino acids, particularly Glu, consistent with increased nitrogen uptake and assimilation. Moreover, the treated cultures showed higher levels of the nitrogen-assimilation enzymes glutamine synthetase and glutamine-2-oxoglutarate-aminotransferase. Finally, 13C acetate labeling showed accumulation of 13C in amino acids. Therefore, in Chlamydomonas cells, inhibition of TOR induced increases in the uptake of nitrogen (preferentially as ammonium) from the medium, and in the assimilation of this nitrogen in amino acid biosynthesis.
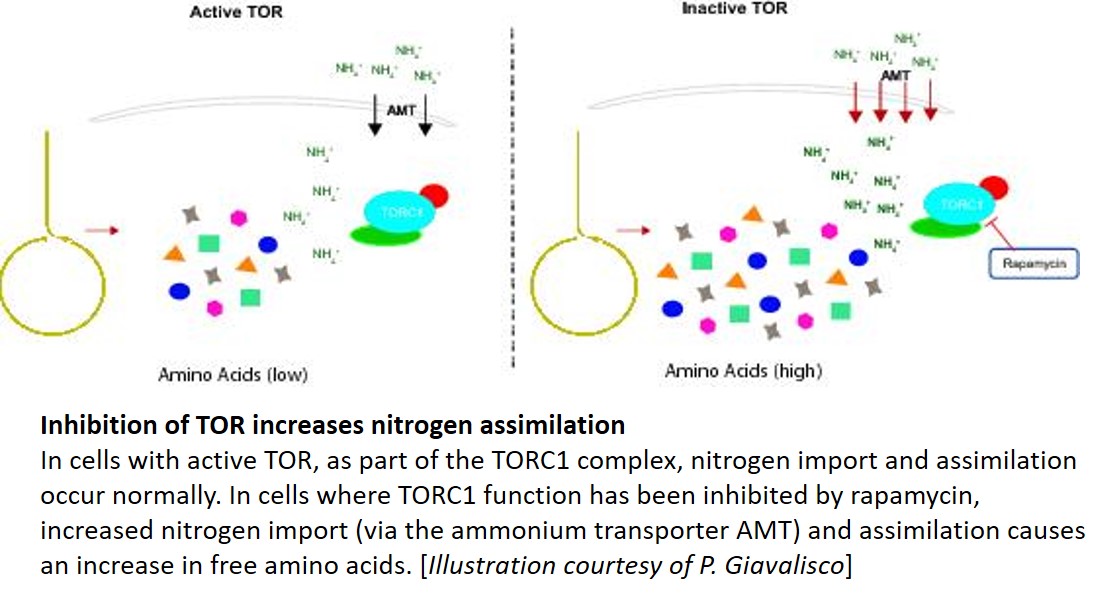
This study gave an unexpected answer to the outstanding question of what is responsible for the accumulation of free amino acids in response to inhibition of TOR. Going forward, identification of the exact targets of TOR in regulating nitrogen import and assimilation remains an interesting problem for future research. Given that TOR inhibition decreases translation, the utility (if any) to the cell of having stockpiles of amino acids also remains unclear. Therefore, this study opens several tempting avenues for investigation, with possible long-term implications for nitrogen use efficiency in crop plants. Looks like we have many more lessons to learn from the green algae.
REFERENCES
Dobrenel, T., Caldana, C., Hanson, J., Robaglia, C., Vincentz, M., Veit, B., and Meyer, C. (2016). TOR Signaling and Nutrient Sensing. Annu. Rev. Plant Biol. 67, 261-285.
Mubeen, U., Jüppner, J., Alpers, J., Hincha, D.K., and Giavalisco, P. (2018). Target of Rapamycin Inhibition in Chlamydomonas reinhardtii Triggers de-novo Amino Acid Synthesis by Enhancing Nitrogen Assimilation. Plant Cell 10.1105/tpc.18.00159.