Handling the Heat – Methylome Variation Underlying Heat Tolerance in Cotton
Plants must accommodate many stresses in order to grow and reproduce normally. Heat stress in particular can negatively affect anther development in flowering plants, leading to male-sterile flowers that produce indehiscent anthers and sterile pollen, and consequently are not capable of sexual reproduction. Increasing global temperatures may lead to decreased yield in cotton, wheat, corn, and rice, intensifying the need to understand the mechanisms underlying heat tolerance in crops. Proper anther and pollen development involves a complex network of transcription factors and small RNAs that result in the reconfiguration of chromatin marks and DNA methylation (reviewed in Kawashima and Berger 2014). Chromatin and the epigenome are uniquely labile throughout plant development, possibly providing a mechanism for plants to adapt to stresses such as heat (Lämke and Bäurle, 2017).
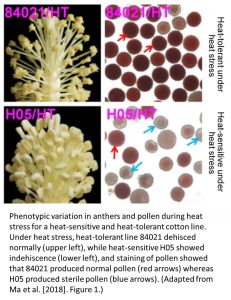 Ma et al. (2018) explore this phenomenon of heat stress using two lines of cotton that differ in their ability to tolerate high temperatures and still produce fertile anthers and pollen (see Figure). They previously discovered that a heat-tolerant (HT) cotton line displays an increase in the overall level of genome-wide DNA methylation when heat-stressed, compared to a heat-sensitive (HS) line that leads to anther indehiscence and pollen sterility when heat-stressed (Min et al. 2014). These methylation differences were previously proposed to underlie the differential expression of genes involved in the starch, auxin, and sugar metabolic pathways that are crucial for proper pollen development. To explore how variation in DNA methylation between these two cotton lines may impact on their ability to tolerate heat stress by modulating these pathways, they performed bisulfite-treated DNA sequencing on tissue from several stages of anther development. By comparing the HS and HT lines under normal conditions and heat stress, they show that DNA methylation in the CHH context (where H=A,T, or G) in the HT line at high temperature is significantly increased uniformly across all chromosomes.
Ma et al. (2018) explore this phenomenon of heat stress using two lines of cotton that differ in their ability to tolerate high temperatures and still produce fertile anthers and pollen (see Figure). They previously discovered that a heat-tolerant (HT) cotton line displays an increase in the overall level of genome-wide DNA methylation when heat-stressed, compared to a heat-sensitive (HS) line that leads to anther indehiscence and pollen sterility when heat-stressed (Min et al. 2014). These methylation differences were previously proposed to underlie the differential expression of genes involved in the starch, auxin, and sugar metabolic pathways that are crucial for proper pollen development. To explore how variation in DNA methylation between these two cotton lines may impact on their ability to tolerate heat stress by modulating these pathways, they performed bisulfite-treated DNA sequencing on tissue from several stages of anther development. By comparing the HS and HT lines under normal conditions and heat stress, they show that DNA methylation in the CHH context (where H=A,T, or G) in the HT line at high temperature is significantly increased uniformly across all chromosomes.
DNA methylation in most plants is controlled by several mechanisms, one of which is the RNA-directed DNA Methylation (RdDM) pathway that produces small RNAs with various functions. CHH methylation is highly associated with repetitive DNA in plants, and can be regulated by the production of predominantly 24 nucleotide (nt) small RNAs in the RdDM pathway. As hypothesized, the HT line contains an increased abundance of 24 nt small RNAs that correlates highly with the increased CHH methylation levels. A handful of genes involved in the RdDM pathway show increased gene expression in the HT line under heat stress compared to the HS line, possibly contributing to genome-wide hypermethylation and the suppression of repetitive elements. Chemical inhibition of DNA methylation using Zebularine showed increased expression of these potentially deleterious repetitive elements, as well as genes involved in amylase activity. An increase in amylase activity under heat stress led to the over-production and accumulation of sugars in the anther, possibly linking anther sterility to genome-wide decreases in DNA methylation levels. These results highlight the complexity of identifying mechanisms underlying stress tolerance in plants, but importantly point to the need for integrated analysis of chromatin modifications, DNA methylation, and developmental biology in the context of plant physiology.
REFERENCES
Kawashima, T., and Berger, F. (2014). Epigenetic Reprogramming in Plant Sexual Reproduction. Nat. Rev. Genet. 15: 613–24.
Lämke, J., and Bäurle, I. (2017). Epigenetic and Chromatin-Based Mechanisms in Environmental Stress Adaptation and Stress Memory in Plants. Genome Biol. 18: 124.
Ma, Y. et al. (2018). Disrupted Genome Methylation in Response to High Temperature Has Distinct Affects on Microspore Abortion and Anther Indehiscence. Plant Cell 30: 1387–1403.
Min, L., Li, Y., Hu, Q., Zhu, L., Gao, W., Wu, Y., Ding, Y., Liu, S., Yang, X., and Zhang, X. (2014). Sugar and Auxin Signaling Pathways Respond to High-Temperature Stress during Anther Development as Revealed by Transcript Profiling Analysis in Cotton. Plant Physiol. 164: 1293–1308.