Genomic variation in 3,010 diverse accessions of Asian cultivated rice (Nature)
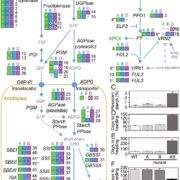
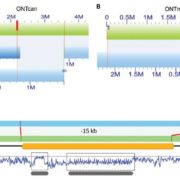
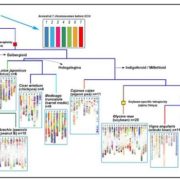
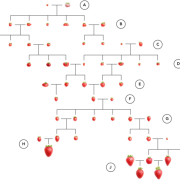
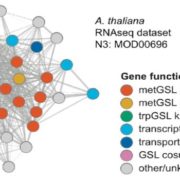
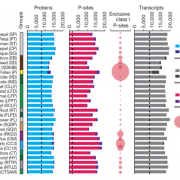
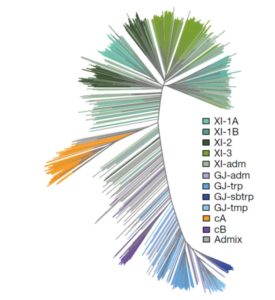 Wang et al. analyse data from the 3000 rice (Oryza sativa) genome (3K-RG) project, identifying “29 million single nucleotide polymorphisms, 2.4 million small indels and over 90,000 structural variations”. The data of course strongly support the two major rice types (Indica and Japonica), but also reveal a total of nine subpopulations. The authors use these data to assemble a rice “pan- genome” comprising all known rice genes. Their data also support multiple different domestication events. Finally, the authors note many genomic structural variations between the Indica and Japonica types which may contribute to their frequently observed hybrid sterility. (Summary by Mary Williams) Nature 10.1038/s41586-018-0063-9
Wang et al. analyse data from the 3000 rice (Oryza sativa) genome (3K-RG) project, identifying “29 million single nucleotide polymorphisms, 2.4 million small indels and over 90,000 structural variations”. The data of course strongly support the two major rice types (Indica and Japonica), but also reveal a total of nine subpopulations. The authors use these data to assemble a rice “pan- genome” comprising all known rice genes. Their data also support multiple different domestication events. Finally, the authors note many genomic structural variations between the Indica and Japonica types which may contribute to their frequently observed hybrid sterility. (Summary by Mary Williams) Nature 10.1038/s41586-018-0063-9