Functional and evolutionary analyses of Cullin1 proteins involved in S-RNase-Based Self-Incompatibility
Sun et al. explore the involvement of CUL1 proteins in S-RNase-based self-incompatibility.
By Linhan Suna and Teh-hui Kaoab
aIntercollege Graduate Degree Program in Plant Biology, The Pennsylvania State University, University Park, Pennsylvania 16802
bDepartment of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania 16802
Background: For any organism, inbreeding reduces fitness of the progeny. To prevent inbreeding, plants have adopted a reproductive strategy, self-incompatibility, to allow pistils to reject genetically identical self-pollen and accept nonself pollen for fertilization. In eudicot families, such as Solanaceae, Plantaginaceae, and Rosaceae, the pistil uses a cytotoxic protein, S-RNase, to inhibit growth of self-pollen tubes. Nonself pollen overcomes S-RNase cytotoxicity using a suite of S-locus F-box (SLF) proteins to mediate degradation of nonself S-RNases. Each SLF is a component of a protein complex, SCFSLF. In Petunia inflata (Solanaceae), this complex contains two pollen-specific components, PiSSK1 and PiCUL1-P.
Question: We wished to determine whether PiCUL1-P is essential for pollen to detoxify nonself S-RNases during cross-compatible pollination, and whether PiCUL1-P functions specifically in self-incompatibility. We also wanted to examine the identity of CUL1 proteins potentially involved in S-RNase-based self-incompatibility in other eudicot families.
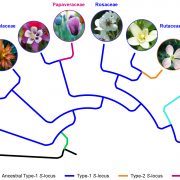
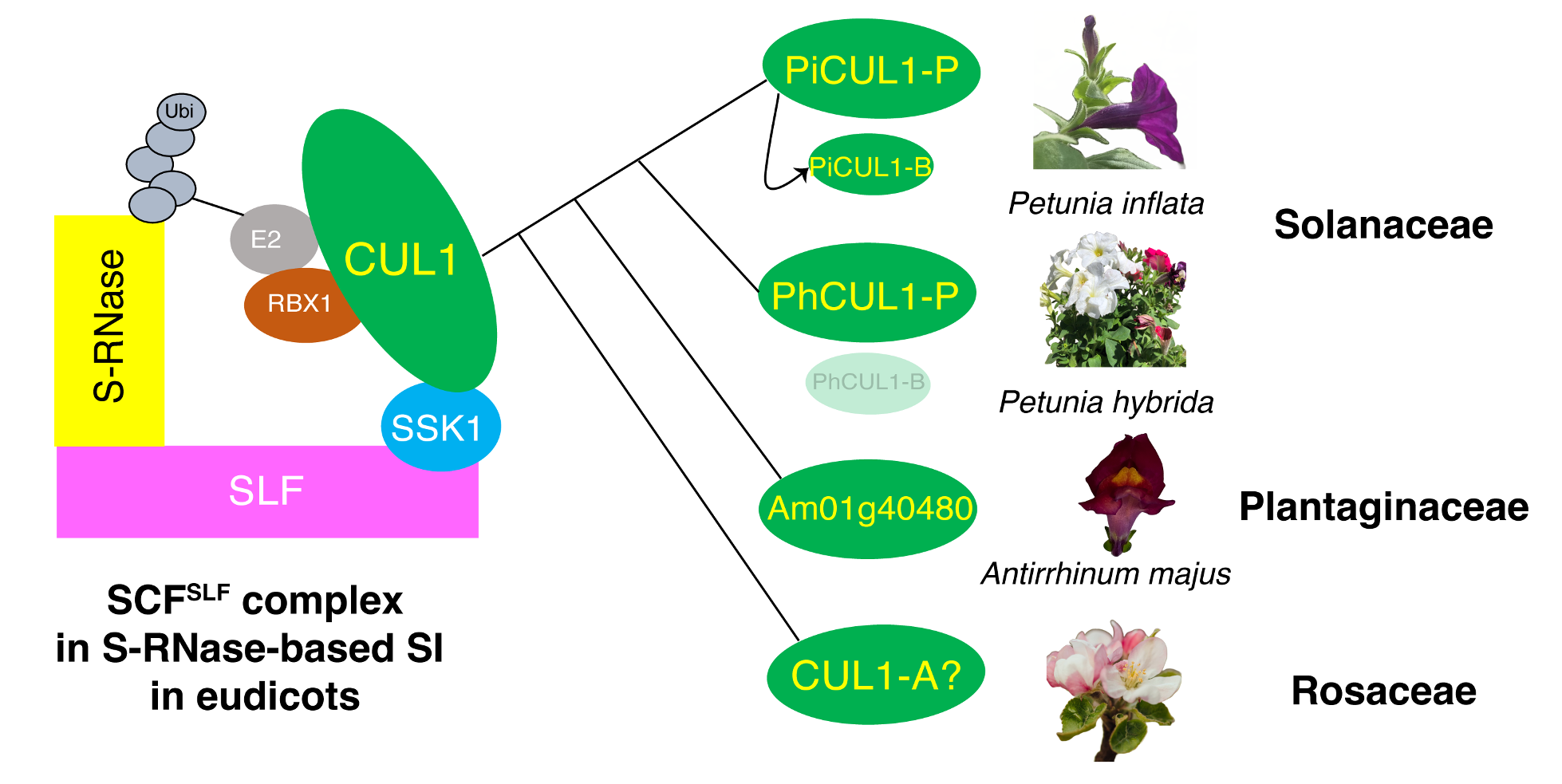 Findings: Using CRISPR/Cas9-mediated gene knockout, we found that, in Petunia inflata, pollen lacking functional PiCUL1-P could still detoxify nonself S-RNases during cross-compatible pollination, whereas pollen lacking both PiCUL1-P and another CUL1, PiCUL1-B, could not, suggesting functional redundancy of these two CUL1 proteins. In Petunia hybrida, we found a transposable element inserted in the promoter region of CUL1-B, which might result in drastic reduction of its transcript level, rendering PhCUL1-P essential for cross-compatibility. We did not find CUL1-B in the other Solanaceae species examined. Analyses of CUL1 evolution in eudicots revealed that all CUL1s involved, or potentially involved, in self-incompatibility possibly share a common ancestor.
Findings: Using CRISPR/Cas9-mediated gene knockout, we found that, in Petunia inflata, pollen lacking functional PiCUL1-P could still detoxify nonself S-RNases during cross-compatible pollination, whereas pollen lacking both PiCUL1-P and another CUL1, PiCUL1-B, could not, suggesting functional redundancy of these two CUL1 proteins. In Petunia hybrida, we found a transposable element inserted in the promoter region of CUL1-B, which might result in drastic reduction of its transcript level, rendering PhCUL1-P essential for cross-compatibility. We did not find CUL1-B in the other Solanaceae species examined. Analyses of CUL1 evolution in eudicots revealed that all CUL1s involved, or potentially involved, in self-incompatibility possibly share a common ancestor.
Next steps: At the biochemical level, we want to determine the amino acids of Petunia CUL1-P and CUL1-B that are responsible for their specific interactions with SSK1 to form SCFSLF complexes. At the evolutionary level, we want to investigate whether the CUL1 and SSK1 components of SCFSLF complexes might have co-evolved in eudicots.
Linhan Sun, Shiyun Cao, Ning Zheng, and Teh-hui Kao. (2023). Analyses of Cullin1 homologs reveal functional redundancy in S-RNase-based self-incompatibility and evolutionary relationships in eudicots https://doi.org/10.1093/plcell/koac357