Frequent Centromere Shifts in a Crucifer Clade
Mandáková et al. investigate genome evolution and frequent centromere repositioning in the tribe Arabideae The Plant Cell (2020). https://doi.org/10.1105/tpc.19.00557
By Martin A. Lysak, CEITEC, Masaryk University, Brno 625 00, Czech Republic.
Background: The centromere is a chromosomal structure that ensures sister-chromatid cohesion and regular chromosome segregation during mitosis and meiosis. Despite their crucial importance, the physical position of centromeres may change without apparent chromosomal rearrangements corrupting chromosomal collinearity. These evolutionary new centromeres (ENCs) have repeatedly been documented in yeast, flies (Drosophila) and vertebrates (e.g., in equids and primates), and less frequently in plants. Triggers, mechanism and evolutionary consequences of ENC formation in diverse eukaryotic phyla are far from being understood.
Question: The unexpected finding of centromere relocation despite the conserved chromosomal collinearity, previously identified in the sequenced Alpine rockcress (Arabis alpina) genome, made us wonder whether the phenomenon is a common feature underlying genome evolution in the whole Arabideae clade from the mustard family (Brassicaceae). We analyzed this by comparative chromosome painting and using the identified centromere-associated tandem repeats.
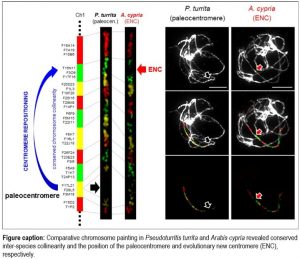 Findings: We found that the intra-tribal cladogenesis in Arabideae (c. 550 species in 18 genera) was marked by a high frequency of ENCs on five out of the eight homoeologous chromosomes in the crown-group genera, but not in the most ancestral Pseudoturritis genome. The high centromere mobility was contrasted by the absence of gross chromosomal rearrangements. While chromosomal localization of ENCs does not reflect the phylogenetic relationship of the Arabideae subclades, centromere repositioning is proposed as the key mechanism differentiating overall conserved homoeologous chromosomes across the crown-group subclades. Our study documented frequent centromere repositioning in the largest species set within a single monophyletic plant clade.
Findings: We found that the intra-tribal cladogenesis in Arabideae (c. 550 species in 18 genera) was marked by a high frequency of ENCs on five out of the eight homoeologous chromosomes in the crown-group genera, but not in the most ancestral Pseudoturritis genome. The high centromere mobility was contrasted by the absence of gross chromosomal rearrangements. While chromosomal localization of ENCs does not reflect the phylogenetic relationship of the Arabideae subclades, centromere repositioning is proposed as the key mechanism differentiating overall conserved homoeologous chromosomes across the crown-group subclades. Our study documented frequent centromere repositioning in the largest species set within a single monophyletic plant clade.
Next steps: Researchers aim to understand what triggers centromere repositioning and whether the formation of ENCs may confer adaptive advantage to their carriers. The role of ENCs as a post-zygotic reproductive barrier in Arabideae is testable by analyzing genomes of closely related (sub)species pairs differentiated by centromere repositioning, and their natural or artificial hybrids. Our work shows Arabideae as a suitable model group to analyze the evolutionary significance of centromere repositioning in plant genome evolution.
Terezie Mandáková, Petra Hlousková, Marcus A. Koch and Martin A. Lysak (2020). Genome Evolution in Arabideae was Marked by Frequent Centromere Repositioning. https://doi.org/10.1105/tpc.19.00557
Key words: Centromere; neocentromere; genome evolution; Brassicaceae




