Epigenetic clocks in plants: uncovering DNA methylation decay in aging
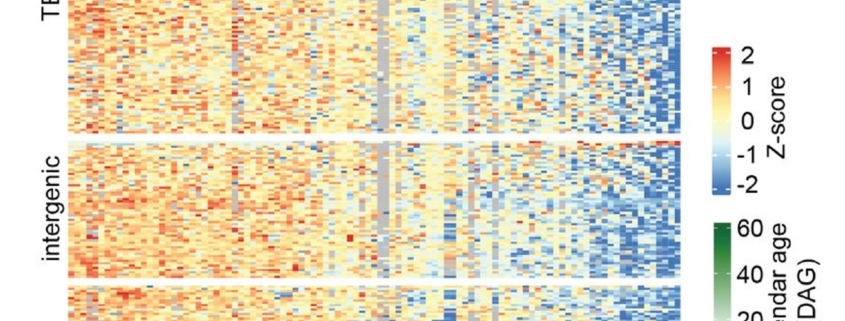
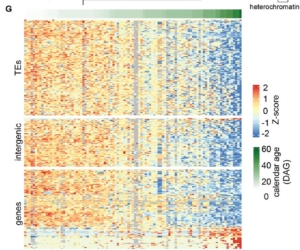 What if plants could teach us about aging? Understanding how and why living organisms age is a fundamental question in biology and medicine. While most research focuses on humans, the model plant Arabidopsis thaliana offers unique insights into how organisms age. In a recent study, Dai et al. analyzed first true leaves at various ages and observed an intriguing pattern: a progressive loss of methylation integrity in heterochromatin regions as the tissues aged. This loss reflects patterns observed in human aging and diseases like cancer. These cumulative methylation losses result in significant differentially methylated regions in senescent leaves, accompanied by an increase in transposable element transcripts. This finding not only highlights the ability of plants to decouple biological and chronological age but also introduces an exciting parallel to the “epigenetic clocks” used to predict biological age in mammals. Even more fascinating is the discovery that new organs are epigenetically young and can age at independent rates from the rest of the organism, suggesting that meristems are “ageless,” producing germline cells and somatic organs that age independently. The study also identifies key regulatory genes, such as TCX5 and TCX6, which act as molecular switches repressing DNA methylation maintenance during aging. Mutants for this mechanism prevent epigenetic aging entirely, opening doors to understanding—and potentially manipulating—aging at a fundamental level. Lastly, but just as importantly, this study highlights that both the organ’s age and the timing of its emergence are critical factors that must be carefully accounted for when sampling! This work emphasizes the tremendous potential of Arabidopsis as a model species in the field of aging research. (Summary by Ileana Tossolini @IleanaDrt @ileanadrt.bsky.social) bioRxiv https://www.biorxiv.org/content/10.1101/2024.11.04.621941v1
What if plants could teach us about aging? Understanding how and why living organisms age is a fundamental question in biology and medicine. While most research focuses on humans, the model plant Arabidopsis thaliana offers unique insights into how organisms age. In a recent study, Dai et al. analyzed first true leaves at various ages and observed an intriguing pattern: a progressive loss of methylation integrity in heterochromatin regions as the tissues aged. This loss reflects patterns observed in human aging and diseases like cancer. These cumulative methylation losses result in significant differentially methylated regions in senescent leaves, accompanied by an increase in transposable element transcripts. This finding not only highlights the ability of plants to decouple biological and chronological age but also introduces an exciting parallel to the “epigenetic clocks” used to predict biological age in mammals. Even more fascinating is the discovery that new organs are epigenetically young and can age at independent rates from the rest of the organism, suggesting that meristems are “ageless,” producing germline cells and somatic organs that age independently. The study also identifies key regulatory genes, such as TCX5 and TCX6, which act as molecular switches repressing DNA methylation maintenance during aging. Mutants for this mechanism prevent epigenetic aging entirely, opening doors to understanding—and potentially manipulating—aging at a fundamental level. Lastly, but just as importantly, this study highlights that both the organ’s age and the timing of its emergence are critical factors that must be carefully accounted for when sampling! This work emphasizes the tremendous potential of Arabidopsis as a model species in the field of aging research. (Summary by Ileana Tossolini @IleanaDrt @ileanadrt.bsky.social) bioRxiv https://www.biorxiv.org/content/10.1101/2024.11.04.621941v1