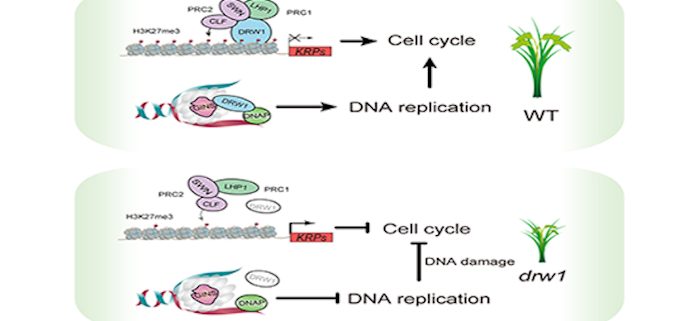
DRW1 interacts with Polycomb complexes but exerts divergent regulatory functions in rice
Zhang et al. explore the mechanisms of epigenetic information transfer in plants.
The Plant Cell
By Pingxian Zhang and Xiaofeng Gu
Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China
Background: Both genetic and epigenetic information must be transferred from mother to daughter cells during cell division. In yeast and mammals, genetic information transfer can be affected by conserved CTF4 (CHROMOSOME TRANSMISSION FIDELITY PROTEIN 4) homologs through influencing DNA helicase activity and error-free DNA damage tolerance in the S phase of the cell cycle. In Arabidopsis thaliana, the homolog of yeast CTF4 (EOL1) not only participates in the DNA helicase complex, but also promotes the recruitment of LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) to sustain histone 3 lysine 27 trimethylation (H3K27me3) methylation levels in dividing cells. The mechanisms of epigenetic information transfer have also been characterized in animals; these processes are less well understood in plants, especially for monocot rice.
Question: We wanted to know whether rice and Arabidopsis homologs of yeast CTF4 are conserved, whether they use common cellular machinery to accomplish similar or divergent functional outcomes, whether they can interact with Polycomb complexes, and how they respond to DNA damage.
Findings: We characterized of a dwarf rice mutant (drw1) deficient for yeast CTF4. We discovered that CTF4 orthologs in plants use common cellular machinery, yet accomplish divergent functional outcomes. Specifically, drw1 exhibited no flowering-related phenotypes (as in the orthologous Arabidopsis eol1 mutant), but displayed cell-cycle arrest and DNA-damage responses. Mechanistically, we demonstrate that DRW1 sustains normal cell-cycle progression by modulating the expression of cell-cycle inhibitors KIP-RELATED PROTEIN 1 (KRP1) and KRP5, and show that these effects are mediated by DRW1 binding their promoters and increasing H3K27me3 levels. Thus, although CTF4 orthologs EOL1 in Arabidopsis and DRW1 in rice are both expressed uniquely in dividing cells, commonly interact with several Polycomb complex subunits, and promote H3K27me3 deposition, we now know that their regulatory functions diverged substantially during plant evolution.
Next steps: We have experimentally determined the specific targets of CTF4/EOL1/DRW1 proteins, and characterized their protein–protein interaction partners and chromatin/epigenetic effects in Arabidopsis and rice. In future, examining the specific binding target genes of CTF4/EOL1/DRW1 family proteins in diverse species will help clarify which biological effects are lineage- or even species-specific.
Pingxian Zhang, Chunmei Zhu, Yuke Geng, Yifang Wang, Ying Yang, Qing Liu, Weijun Guo, Sadaruddin Chachar, Adeel Riaz, Shuangyong Yan, Liwen Yang, Keke Yi, Changyin Wu, Xiaofeng Gu. (2021). Rice and Arabidopsis homologs of yeast CHROMOSOME TRANSMISSION FIDELITY PROTEIN 4 commonly interact with Polycomb complexes but exert divergent regulatory functions. Plant Cell.