Double trouble: The Solanum pan-genome shows gene duplication complicates predictability
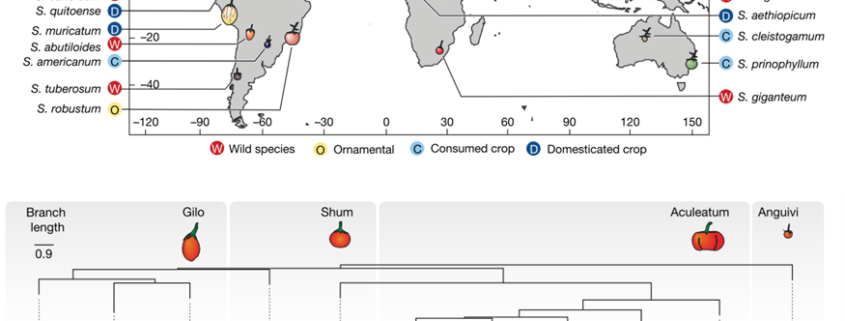
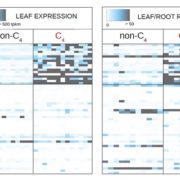
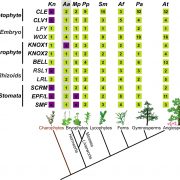
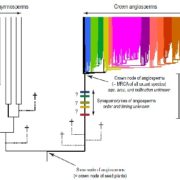
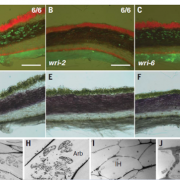
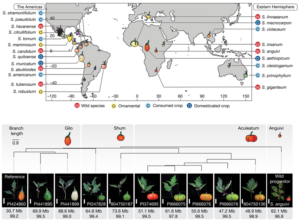 A pan-genome has been assembled for the Solanum genus, which contains many diverse and economically important crops including potato, tomato, and African eggplant. Genomes were assembled for 22 species, and genes were predicted based on previous reference genomes and from RNA sequences across multiple tissues. Approximately 60% of genes were the core genome (found in all species), and while synteny was mostly conserved across the species, large scale inversions and translocations were seen within subclades. While gene content was largely consistent between species, genome sizes ranged from 0.7-2.5 Gb mainly due to retrotransposon activity, but also from gene duplications. Over half a million duplications were found, both whole genome and single-gene, and paralogues disproportionally evolved to maintain normal gene dosage. This was greatly achieved through conservation of the cis-regulatory sequences compared to the coding portion of genes through evolution. Paralogues with similar expression patterns may be partially or fully redundant, compensatory, or pseudogenized, which may mean related genes in another species have different functions and effects on plant phenotypes. This complicates genetic engineering for crop improvement, as the effect on phenotype in one species may not reliably predict the outcome in others. However, with the continual establishment of lineage-specific pan-genomes, combined with advances in machine learning models, enhancement in the prediction accuracy is anticipated, paving the way for more precise and efficient crop improvement strategies. (Summary by Ciara O’Brien @ciara.obrien.bsky.social) Nature 10.1038/s41586-025-08619-6
A pan-genome has been assembled for the Solanum genus, which contains many diverse and economically important crops including potato, tomato, and African eggplant. Genomes were assembled for 22 species, and genes were predicted based on previous reference genomes and from RNA sequences across multiple tissues. Approximately 60% of genes were the core genome (found in all species), and while synteny was mostly conserved across the species, large scale inversions and translocations were seen within subclades. While gene content was largely consistent between species, genome sizes ranged from 0.7-2.5 Gb mainly due to retrotransposon activity, but also from gene duplications. Over half a million duplications were found, both whole genome and single-gene, and paralogues disproportionally evolved to maintain normal gene dosage. This was greatly achieved through conservation of the cis-regulatory sequences compared to the coding portion of genes through evolution. Paralogues with similar expression patterns may be partially or fully redundant, compensatory, or pseudogenized, which may mean related genes in another species have different functions and effects on plant phenotypes. This complicates genetic engineering for crop improvement, as the effect on phenotype in one species may not reliably predict the outcome in others. However, with the continual establishment of lineage-specific pan-genomes, combined with advances in machine learning models, enhancement in the prediction accuracy is anticipated, paving the way for more precise and efficient crop improvement strategies. (Summary by Ciara O’Brien @ciara.obrien.bsky.social) Nature 10.1038/s41586-025-08619-6