Do all roads lead to 2-phenylethanol in Populus?
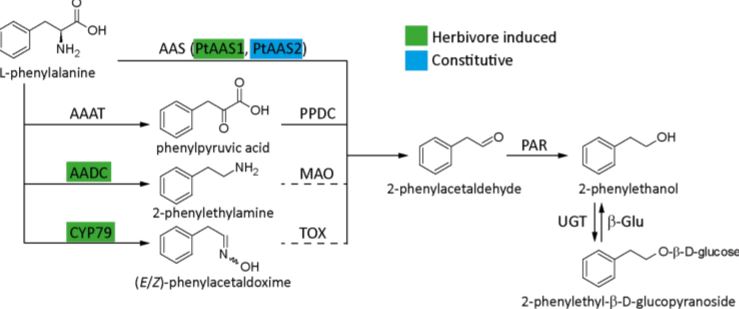
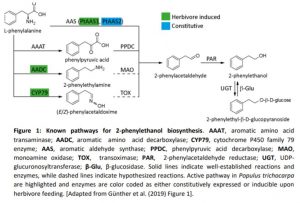 Plants emit scented volatile compounds to attract pollinators, or alternatively, as direct or indirect plant defense mechanisms. Direct defense involves compounds that are toxic, whereas an indirect defense uses volatiles to attract parasitoids of insect herbivores (Unsicker et al., 2009; Maag et al., 2015). Therefore, volatile compounds are undoubtedly important for plants, in addition they also have commercial value; and for both reasons their biosynthetic pathways have become well studied. However, multiple pathways producing the same compound are used in different species and in a few cases multiple pathways are present in one species. This raises the question: what pathway(s) is used in planta? The amino acid derived 2-phenylethanol, a volatile compound with a rose-like scent, is one such compound. It is produced by over 50 plant species and is involved in pollinator attraction and both direct and indirect defense, and there are currently four known possible biosynthetic pathways, all starting from phenylalanine (Figure 1).
Plants emit scented volatile compounds to attract pollinators, or alternatively, as direct or indirect plant defense mechanisms. Direct defense involves compounds that are toxic, whereas an indirect defense uses volatiles to attract parasitoids of insect herbivores (Unsicker et al., 2009; Maag et al., 2015). Therefore, volatile compounds are undoubtedly important for plants, in addition they also have commercial value; and for both reasons their biosynthetic pathways have become well studied. However, multiple pathways producing the same compound are used in different species and in a few cases multiple pathways are present in one species. This raises the question: what pathway(s) is used in planta? The amino acid derived 2-phenylethanol, a volatile compound with a rose-like scent, is one such compound. It is produced by over 50 plant species and is involved in pollinator attraction and both direct and indirect defense, and there are currently four known possible biosynthetic pathways, all starting from phenylalanine (Figure 1).
In this issue of Plant Physiology, Günther et al. (2019) investigate which pathway(s) the model poplar species Populus trichocarpa uses for 2-phenylethanol biosynthesis. It is one of the major volatile compounds produced after herbivore feeding, along with 2-phenylethylamine and its nonvolatile glycoside, 2-phenylethyl-b-D-glucopyranoside. A previous study showed that the CYP79 pathway (Figure 1) contributes to 2-phenylethanol biosynthesis, but also indicated that more than one pathway might be used (Irmisch et al., 2013; Irmisch et al., 2014). To further investigate which pathway is used, the authors performed a combination of phytochemical analysis, biochemical characterization, and RNAi-mediated downregulation of enzymes with potential involvement in 2-phenylethanol biosynthesis.
Aromatic aldehyde synthase 1 (PtAAS1) and PtAAS2 were shown to catalyze 2-phenylacetaldehyde formation from phenylalanine, with PtAAS1 being specific towards this one substrate, while PtAAS2 accepted other aromatic amino acids and produced their respective aldehydes. The aldehyde is further converted into 2-phenylethanol by 2-phenylacetaldehyde reductase 1 (PtPAR1) and PtPAR2, although neither enzyme seems to be specific to this pathway alone as they show broad substrate specificity. The enzymes phenylpyruvic acid decarboxylase 1 (PtAADC1) and PtAADC2, although closely related to the AAS enzymes, produce 2-phenylethylamine instead. Both also accept other aromatic amino acids as substrates and produce their respective amines. The different activities of AAS and AADC enzymes, producing either the corresponding aldehyde or an amine from an amino acid, was traced to one amino acid change in the active site. This was already previously shown by another group (Torrens-Spence et al., 2013). Interestingly, PtAAS1 and PtAADC1 are clustered on chromosome 13 and a similar clustering of an AADC/AAS pairs was shown for 10 other plant species, suggesting recent tandem gene duplication events.
As P. trichocarpa is not amenable to genetic transformation, a combination of overexpression of PtAAS1 and PtAADC1 in Nicotiana benthamiana, and RNAi-mediated downregulation of AADC1 in Populus x canescens was done, confirming the observed in vitro activities. Surprisingly, the amino acid, that decides whether an enzyme is producing an aldehyde (AAS) or an amine (AADC) is altered in the PtAADC2 ortholog of Populus x canescens and converts it to an AAS. However, neither this ortholog nor the PtAAS1 ortholog are expressed in Populus x canescens. RNAi knock down of the PtAADC1 ortholog reduced 2-phenylethylamine and 2-phenylethyl-b-D-glucopyranoside production, while having no effect on 2-phenylethanol emission. The authors suggest that PtAADC1 forms a metabolon with downstream enzymes leading to production of 2-phenylethyl-b-D-glucopyranoside. The implication is that 2-phenylethanol is likely produced by three pathways in P. trichocarpa, which are either constitutively expressed or are induced upon herbivory.
This study illustrates that more than one pathway can be used in plants to produce a single compound and that biosynthesis is often not linear, but rather a complex network. It also cautions us not to make assumptions about the function or regulation of pathways in two species. This research particularly illustrates the fact that pathways can either be non-functional, have mutations that change functionality, or have enzymes that are not actively expressed. Yet the most significant take away message is that a change of one amino acid can drastically alter the functionality of an enzyme. While the importance of this amino acid was previously known (Torrens-Spence et al., 2013), Günther et al. (2019) showed that this amino acid can differ between closely related species.
Figure 1: Known pathways for 2-phenylethanol biosynthesis. AAAT, aromatic amino acid transaminase; AADC, aromatic amino acid decarboxylase; CYP79, cytochrome P450 family 79 enzyme; AAS, aromatic aldehyde synthase; PPDC, phenylpyruvic acid decarboxylase; MAO, monoamine oxidase; TOX, transoximase; PAR, 2-phenylacetaldehyde reductase; UGT, UDP-glucuronosyltransferase; β-Glu, β-glucosidase. Solid lines indicate well-established reactions and enzymes, while dashed lines indicate hypothesized reactions. Active pathway in Populus trichocarpa are highlighted and enzymes are color coded as either constitutively expressed or inducible upon herbivore feeding. [Adapted from Günther et al. (2019) Figure 1].
LITERATURE CITED
Günther J, Lackus ND, Schmidt A, Huber M, Stödler H-J, Reichelt M, Gershenzon J, Köllner TG (2019) Separate pathways contribute to the herbivore-induced formation of 2-phenylethanol in poplar. Plant Physiology: pp.00059.02019 https://doi.org/10.1104/pp.19.00059
Irmisch S, Clavijo McCormick A, Boeckler GA, Schmidt A, Reichelt M, Schneider B, Block K, Schnitzler J-P, Gershenzon J, Unsicker SB, Köllner TG (2013) Two Herbivore-Induced Cytochrome P450 Enzymes CYP79D6 and CYP79D7 Catalyze the Formation of Volatile Aldoximes Involved in Poplar Defense. The Plant Cell 25: 4737-4754
Irmisch S, Clavijo McCormick A, Günther J, Schmidt A, Boeckler GA, Gershenzon J, Unsicker SB, Köllner TG (2014) Herbivore-induced poplar cytochrome P450 enzymes of the CYP71 family convert aldoximes to nitriles which repel a generalist caterpillar. The Plant Journal 80: 1095-1107
Maag D, Erb M, Kollner TG, Gershenzon J (2015) Defensive weapons and defense signals in plants: some metabolites serve both roles. Bioessays 37: 167-174
Torrens-Spence MP, Liu P, Ding H, Harich K, Gillaspy G, Li J (2013) Biochemical Evaluation of the Decarboxylation and Decarboxylation-Deamination Activities of Plant Aromatic Amino Acid Decarboxylases. Journal of Biological Chemistry 288: 2376-2387
Unsicker SB, Kunert G, Gershenzon J (2009) Protective perfumes: the role of vegetative volatiles in plant defense against herbivores. Current Opinion in Plant Biology 12: 479-485