Die Another Way: An EDS1-SAG101 Complex Mediates TNL Immunity in Solanaceous Plants
Disease resistance (R) receptors detect pathogen effector-mediated host manipulation and induce effector-triggered immunity (ETI) that is often associated with programmed cell death. R proteins are generally conserved across plants, consisting of nucleotide-binding site and leucine rich repeat (NBS-LRR) domains alongside a variable N-terminal coiled-coil (CC) or Toll-like Interleukin receptor (TIR) domain; referred to as CNL or TNL receptors, respectively (Adachi et al., 2019). Genetic dissection of TNL immunity in Arabidopsis thaliana has identified a strict dependency on the lipase-like protein ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), which forms heteromeric complexes with sequence-related PHYTOALEXIN DEFICIENT4 (PAD4) or SENESCENCE ASSOCIATED GENE101 (SAG101) that vary in their ability to contribute to immune signaling (Cui et al., 2015).
Two complementary studies offer new insights into the evolutionary history and functional diversity of EDS1 family genes in angiosperms. Gantner et al. (2019) take a functional genomic approach to dissect EDS1-dependent TNL immunity through a combination of CRISPR/Cas9 gene editing and Agrobacterium-mediated transient expression-based rescue experiments in the model angiosperm Nicotiana benthamiana. By contrast, Lapin et al. (2019) begin with an evolutionary perspective, interrogating 52 green plant genomes for EDS1 family member homologs that the authors functionally investigate in Arabidopsis (family Brasicaceae) and Nicotiana (family Solanacaea).
Phylogenetic analysis of EDS1 family genes revealed broad conservation of EDS1 and PAD4 orthologs across seed plants (gymnosperms and terrestrial angiosperms), whereas SAG101 homologs appear to be absent from gymnosperms and angiosperm lineages lacking TNL receptors (monocots, dicots belonging to Caryophyllales) (Lapin et al., 2019). This analysis, alongside that of Gantner et al. (2019), identified candidate EDS1, PAD4 and SAG101a/b (gene duplication) homologs in the Solanaceaous plants N. benthamiana and Solanum lycopersicum (tomato). Using complementary sets of protein-protein interaction assays, both groups identified strong conservation of intra-species heterodimer complex formation between EDS1 and PAD4/SAG101 homologs within Arabidopsis or tomato (Gantner et al., 2019; Lapin et al., 2019). In addition, expanded yeast two-hybrid analyses revealed occasional inter-species EDS1-PAD4 interactions in angiosperm models other than Arabidopsis (Lapin et al., 2019).
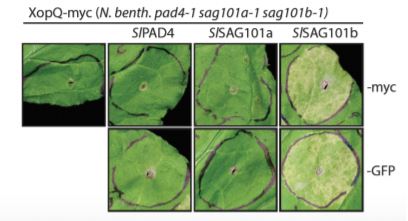
 CRISPR/Cas-generated eds1 family mutants, tested alone or in combination, revealed a conserved role for NbEDS1 and, intriguingly, a specific requirement for NbSAG101b in TNL (Roq1-mediated) immunity/cell death activated by the Xanthomonas campestris pv. vesicatoria effector XopQ (Gantner et al., 2019; Lapin et al., 2019). This is in contrast to Arabidopsis, where the EDS1-PAD4 complex predominantly contributes to TNL-based ETI (Wagner et al., 2013). A generalizable role for NbEDS1-NbSAG101b in TNL immunity was further supported by its requirement for cell death signaled through additional truncated (autoactivated) or full-length TNL receptors (DM2hTIR, RPS4TIR and N; Gantner et al., 2019). Cross species complex transferability assays revealed that SlEDS1-SlPAD4 co-expression rescue Arabidopsis eds1/pad4/sag101 mutants defective in RPP4 or RPP2-mediated TNL immunity to avirulent Hyaloperonospora arabidopsidis, whereas AtEDS1-AtPAD4/AtSAG101 could not rescue Nicotiana eds1 family mutants. Gantner and colleagues speculated that an additional Arabidopsis factor was lacking, which Lapin et al. (2019) reveal to be the AtNRG1 helper NLR. This indicated a requirement for matching (co-evolved) signaling proteins to function in TNL immunity and host cell death.
CRISPR/Cas-generated eds1 family mutants, tested alone or in combination, revealed a conserved role for NbEDS1 and, intriguingly, a specific requirement for NbSAG101b in TNL (Roq1-mediated) immunity/cell death activated by the Xanthomonas campestris pv. vesicatoria effector XopQ (Gantner et al., 2019; Lapin et al., 2019). This is in contrast to Arabidopsis, where the EDS1-PAD4 complex predominantly contributes to TNL-based ETI (Wagner et al., 2013). A generalizable role for NbEDS1-NbSAG101b in TNL immunity was further supported by its requirement for cell death signaled through additional truncated (autoactivated) or full-length TNL receptors (DM2hTIR, RPS4TIR and N; Gantner et al., 2019). Cross species complex transferability assays revealed that SlEDS1-SlPAD4 co-expression rescue Arabidopsis eds1/pad4/sag101 mutants defective in RPP4 or RPP2-mediated TNL immunity to avirulent Hyaloperonospora arabidopsidis, whereas AtEDS1-AtPAD4/AtSAG101 could not rescue Nicotiana eds1 family mutants. Gantner and colleagues speculated that an additional Arabidopsis factor was lacking, which Lapin et al. (2019) reveal to be the AtNRG1 helper NLR. This indicated a requirement for matching (co-evolved) signaling proteins to function in TNL immunity and host cell death.
 Evolutionarily- and structurally-guided analysis of the EDS1-SAG101 complex interface further delineated protein domains and amino acid residues essential for heterodimer formation and TNL immunity in N. benthamiana. Functional analysis of AtPAD4-AtSAG101 gene chimeras (co-expressed with AtEDS1 and AtNRG1 [Lapin et al., 2019]), or NbSAG101a-NbSAG101b chimeras (Gantner et al., 2019), highlighted the importance of a surface exposed a-helical region located in the conserved EP domain. Together, these studies reveal fine-tuned constraints on EDS1 family complexes that co-evolved to regulate TNL-based immunity in seed plants. Moreover, the contrasting roles of PAD4 and SAG101 in establishing functional heterodimer complexes with EDS1 in different angiosperms further underscores the importance of understanding how evolution has shaped plant immune networks, which may ultimately guide future works aimed at mechanistically unravelling EDS1-mediated immunity.
Evolutionarily- and structurally-guided analysis of the EDS1-SAG101 complex interface further delineated protein domains and amino acid residues essential for heterodimer formation and TNL immunity in N. benthamiana. Functional analysis of AtPAD4-AtSAG101 gene chimeras (co-expressed with AtEDS1 and AtNRG1 [Lapin et al., 2019]), or NbSAG101a-NbSAG101b chimeras (Gantner et al., 2019), highlighted the importance of a surface exposed a-helical region located in the conserved EP domain. Together, these studies reveal fine-tuned constraints on EDS1 family complexes that co-evolved to regulate TNL-based immunity in seed plants. Moreover, the contrasting roles of PAD4 and SAG101 in establishing functional heterodimer complexes with EDS1 in different angiosperms further underscores the importance of understanding how evolution has shaped plant immune networks, which may ultimately guide future works aimed at mechanistically unravelling EDS1-mediated immunity.
Philip Carella
Sainsbury Laboratory
University of Cambridge, Cambridge
REFERENCES
Adachi H, Derevnina L and Kamoun S (2019). NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50: 121-131.
Cui H, Tsuda K and Parker JE (2015). Effector-triggered immunity: from pathogen perception to robust defense. Ann. Rev. Plant Biol. 66: 487-511.
Gantner J, Ordon J, Kretschmer C, Guerois R and Stuttmann (2019). An EDS1-SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana. Plant Cell Published July 2019 DOI: https://doi.org/10.1105/tpc.19.00099.
Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, Beyer A and Parker JE (2019). A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell Published July 2019 DOI: https://doi.org/10.1105/tpc.19.00118.
Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K and Parker JE (2013). Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Micr. 14: 619-630.
Figure legend:
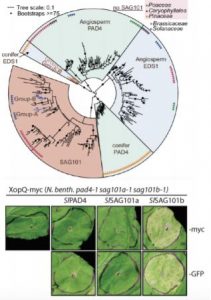
Evolution and functional diversification EDS1-related genes.
Upper panel: Phylogenetic analysis of EDS1 family homologs identified by interrogating 52 green plant genomes reveals evolutionary relationships and distinct EDS1 and PAD4 clades specific to angiosperms (flowering) versus conifers (non-flowering) seed plants, whereas a single SAG101 lineage specific to certain angiosperms is observed (Adapted from Lapin et al., [2019], Figure 1). Lower panel: Functional complementation/rescue of XopQ-triggered cell death in the N. benthamiana pad4/sag101a/sag101b triple mutant background occurs only with expression of SAG101b homologs (tomato SlSAG101b shown) irrespective of protein fusion tags (Adapted from Gantner et al., [2019], Figure 3).