Defining the repertoire of fungal effector proteins deployed during plant infection
Yan et al. explore the changes in gene expression that take place during rice blast disease.
By Xia Yan and Nick Talbot from The Sainsbury Laboratory, UK.
Background: The rice blast fungus Magnaporthe oryzae causes a devastating disease that threatens global rice (Oryza sativa) production, destroying enough rice each year to feed more than 60 million people. Although the disease has been extensively studied, we still do not understand how the fungus is able to invade rice tissue so effectively and how it overwhelms the plant’s defenses.
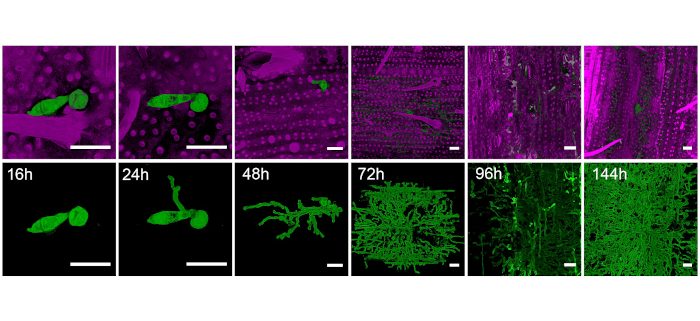
Question: We set out to identify the major changes in gene expression that occur during rice blast disease and to use this information to define the repertoire of fungal effector proteins that are deployed by the fungus during plant infection.
Findings: We discovered that the blast fungus has a much more extensive repertoire of effectors than previously thought and these effectors are expressed at specific times during infection. This mirrors the expression of very large sets of fungal genes encoding proteins associated with secondary metabolism, transporter functions, and cell signalling during each stage of pathogenesis. We found that sequence un-related but structurally conserved effectors are expressed in a co-ordinated manner during infection. Many of these effectors are delivered into plant cells and we developed a sensitive relative fitness assay to show that even an individual effector can contribute to fungal virulence.
Next steps: Two main questions emerge from our study. First, what do all of these effectors do? What are their targets and how do they enable the fungus to colonize rice cells so rapidly? Second, how are effector genes temporally and spatially regulated? What are the transcriptional regulators that ensure they are expressed in such specific patterns?
Xia Yan, Bozeng Tang, Lauren S. Ryder, Dan MacLean, Vincent M. Were, Alice Bisola Eseola, Neftaly Cruz-Mireles, Weibin Ma, Andrew J. Foster, Miriam Osés-Ruiz and Nicholas J. Talbot. (2023). The transcriptional landscape of plant infection by the rice blast fungus Magnaporthe oryzae reveals distinct families of temporally co-regulated and structurally conserved effectors https://doi.org/10.1093/plcell/koad036