Decoding Natural Variation in Chloroplast Size
Lynn GL Richardson
Department of Plant Biology, Michigan State University, East Lansing, MI 48824
Chloroplasts, like their cyanobacterial ancestors, use binary fission to divide and generate new organelles. The origins of the division machinery stem from both the original eukaryotic host cell and cyanobacterial endosymbiont (Chen et al., 2018). During chloroplast division, a contractile ring assembles at each of the outer and inner chloroplast membranes and together coordinate constriction and pinching off of both membranes to create two daughter chloroplasts (Figure 1A). Mutation of individual components of the chloroplast division machinery typically results in larger chloroplasts, with fewer chloroplasts per cell (Chen et al., 2018). Defects in chloroplast division have been linked to abnormal chloroplast movement, effects on photosynthesis, and reduced mesophyll conductance (Austin Li and Webber, 2005; Dutta et al., 2017). In nature, chloroplast number or size can vary between species (Honda et al., 1971) and also between cell types of the same species (Ahmadabadi and Bock, 2012); however, the genes/alleles responsible for this variation are unknown. In this issue of Plant Physiology, Kadirjan-Kalbach and colleagues (2019) explored the molecular mechanisms underlying natural variation in chloroplast size using different accessions of Arabidopsis thaliana.
At the start of chloroplast division, division machinery assembles at the organelle midzone on the surface of the inner membrane, forming the “Z-ring”. Correspondingly, the outer envelope plastid division ring, or “PD-ring”, assembles on the surface of the chloroplast (Figure 1A). DYNAMIN-RELATED PROTEIN 5 (DRP5; also known as ACCUMULATION AND REPLICATION OF CHLOROPLASTS 5, or ARC5) is recruited from the cytosol to the division site and initiates dimerization of the outer membrane proteins, PLASTID DIVISION 1 & 2 (PDV1 & 2, respectively) (Chen et al., 2018) (Figure 1A). In a chain reaction that follows, dimerization of the inner membrane protein ARC6 via interactions with PDV1 & 2 in the intermembrane space induces Z-ring condensation (Figure 1A). The Z-ring is composed of the tubulin-like GTPase ring proteins, FILAMENTING TEMPERATURE-SENSITIVE MUTANT Z 1 and 2 (FtsZ1 and 2). FtsZ2 is important for Z-ring structure, while FtsZ1 is a more dynamic component, allowing for tightening of the Z-ring during division (Figure 1A) (Chen et al., 2018). In Arabidopsis, FtsZ1 is encoded by a single gene (FtsZ1-1), while two versions of FtsZ2 exist, encoded by FtsZ2-1 and FtsZ2-2. FtsZ2-1 and FtsZ2-2 appear to be functionally interchangeable—it is the overall level of FtsZ2 protein that is critical for proper Z-ring function (Schmitz et al., 2009).
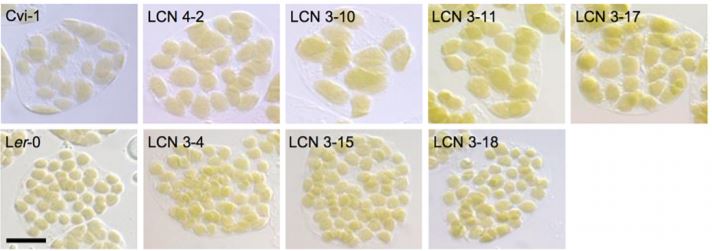
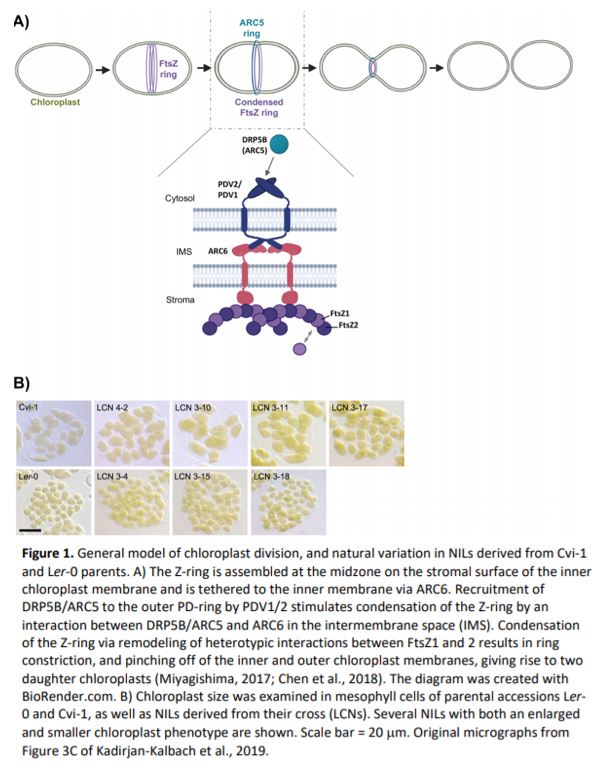 Kadirjan-Kalbach and colleagues (2019) began by taking advantage of existing near-isogenic lines (NILs) originating from parents that exhibit notable differences in chloroplast size. In an initial screen, the chloroplast area per mesophyll cell was measured in 22 Arabidopsis accessions, with the largest difference in chloroplast size occurring between Cvi-1 (large) and Ler-2 or Ler-0 (small). Existing NILs (near isogenic lines; generated by crossing Ler-0 and Cvi-1 parents: LCNs) (Keurentjes et al., 2007) were then analyzed for chloroplast size, revealing 18 LCNs with enlarged chloroplasts similar to Cvi-1 (Figure 1B). These all had Cvi-1 introgressions in chromosome 3, and fine-mapping identified two possible responsible loci, one of which encoded FtsZ2-2 (Kadirjan-Kalbach et al., 2019). Given its known role in chloroplast division in Col-0, the FtsZ2-2 gene from Cvi-1 was sequenced and compared to that of Ler-0. Within Cvi-1, many polymorphisms were identified that distinguished Cvi-1 FtsZ2-2 from that of Ler-0. However, sequencing of several NILs with large chloroplasts pointed toward a SNP in the last exon of FtsZ2-2 that produced a premature stop codon, predicted to encode a protein 18 amino acids shorter than that of Ler-0 FtsZ2-2. A truncated FtsZ2-2 protein was detected in all NILs with large chloroplasts using immunoblotting, which was not due to alternative splicing of the FtsZ2-2 mRNA (Kadirjan-Kalbach et al., 2019).
Kadirjan-Kalbach and colleagues (2019) began by taking advantage of existing near-isogenic lines (NILs) originating from parents that exhibit notable differences in chloroplast size. In an initial screen, the chloroplast area per mesophyll cell was measured in 22 Arabidopsis accessions, with the largest difference in chloroplast size occurring between Cvi-1 (large) and Ler-2 or Ler-0 (small). Existing NILs (near isogenic lines; generated by crossing Ler-0 and Cvi-1 parents: LCNs) (Keurentjes et al., 2007) were then analyzed for chloroplast size, revealing 18 LCNs with enlarged chloroplasts similar to Cvi-1 (Figure 1B). These all had Cvi-1 introgressions in chromosome 3, and fine-mapping identified two possible responsible loci, one of which encoded FtsZ2-2 (Kadirjan-Kalbach et al., 2019). Given its known role in chloroplast division in Col-0, the FtsZ2-2 gene from Cvi-1 was sequenced and compared to that of Ler-0. Within Cvi-1, many polymorphisms were identified that distinguished Cvi-1 FtsZ2-2 from that of Ler-0. However, sequencing of several NILs with large chloroplasts pointed toward a SNP in the last exon of FtsZ2-2 that produced a premature stop codon, predicted to encode a protein 18 amino acids shorter than that of Ler-0 FtsZ2-2. A truncated FtsZ2-2 protein was detected in all NILs with large chloroplasts using immunoblotting, which was not due to alternative splicing of the FtsZ2-2 mRNA (Kadirjan-Kalbach et al., 2019).
Arabidopsis Col-0 ftsZ2-2 mutants have large chloroplasts with fewer per cell (Schmitz et al., 2009). To confirm that the Cvi-1 FtsZ2-2 allele results in larger chloroplasts, it was introduced into the ftsZ2-2 knockout background. Chloroplast size was similar in the ftsZ2-2 mutant with or without the addition of Cvi-FtsZ2-2, while expression of Ler-0 FtsZ2-2 in the ftsz2-2 background restored chloroplast size to that of Col-0. Immunofluorescent labeling of FtsZ2-1 and FtsZ2-2 in both Ler-0 and Cvi-1 showed that in Cvi-1, both Z-ring proteins were found not only at the chloroplast midzone, but also in punctate and short filament structures that were absent from Ler-0 (Kadirjan-Kalbach et al., 2019). This suggested that larger chloroplasts in Cvi-1 may be due to differences in Z-ring organization during plastid division.
The discovery of a unique FtsZ2-2 allele controlling chloroplast size in Arabidopsis Cvi-1 prompted the authors to investigate additional FtsZ2-2 polymorphisms in other Arabidopsis accessions. Using the 1001 Genomes database, the FtsZ2-2 amino acid sequence obtained from 1,135 accessions was compared to that of Col-0. Three accessions had amino acid deletions in FtsZ2-2, including Cvi-0, which was identical in sequence to Cvi-1; and TAD-04, which had a premature stop codon, effectively resulting in a null FtsZ2-2 allele. Both of these accessions had large chloroplasts. A third accession, ANH-1 had an amino acid deletion within the chloroplast targeting signal of FtsZ2-2, which had no effect on the localization of FtsZ2-2 or chloroplast size relative to Ler-0 (Kadirjan-Kalbach et al., 2019). Thirteen accessions contained one or more amino acid polymorphisms in FtsZ2-2. Of these 13, two accessions (Vdm-0 and Sac-0) had unique amino acid substitutions and a quantitative increase in chloroplast size. However, FtsZ2-2 protein levels were much lower in these two accessions compared to Cvi-1 and Ler-0, without differences in transcript levels, suggesting instability of the FtsZ2-2 protein (Kadirjan-Kalbach et al., 2019).
The presence of several unique FtsZ2-2 alleles in different Arabidopsis accessions raised the question of whether they may be under evolutionary selection. Using data available in the 1001 Genomes database, the authors calculated the number of substitutions per non-synonymous site (amino acid change; dN) and the number of substitutions per synonymous site (no amino acid change; dS) for FtsZ1, FtsZ2-1 and FtsZ2-2 in available Arabidopsis accessions. dN/dS gives information about the strength and mode of natural variation for a particular gene and can provide insight into its evolutionary history (Jeffares et al., 2015). A low dN/dS (<1) indicates mutations that cause amino acid changes are not retained, as they may be deleterious to protein folding and/or function (Jeffares et al., 2015). Most protein-coding genes fall into this category. FtsZ1, FtsZ2-1 and FtsZ2-2 all had a dN/dS <1, suggesting negative selection against non-synonymous polymorphisms (Kadirjan-Kalbach et al., 2019). However, the authors point out that the relatively high dN/dS of FtsZ2-2 compared to that of FtsZ1 and FtsZ2-1 may indicate a more relaxed tolerance of FtsZ2-2 for variation.
This study nicely shows the utility in mining natural variation to understand fundamental cell biology processes and builds upon foundational work identifying the critical players involved in chloroplast division in Arabidopsis. Given our extensive use of Arabidopsis as a model plant, especially for cell and molecular biology, similar approaches will undoubtedly be useful in understanding more aspects of plant cell biology. This approach may also help inform our understanding of how chloroplast size is determined in species other than Arabidopsis, including those that show cell type-specific variation in chloroplast size (Ahmadabadi and Bock, 2012).
References
Ahmadabadi M, Bock R (2012) Plastid division and morphology in the genus Peperomia. Biologia Plantarum 56: 301-306
Austin Li J, Webber AN (2005) Photosynthesis in Arabidopsis thaliana mutants with reduced chloroplast number. Photosynth Res 85: 373-384
Chen C, MacCready JS, Ducat DC, Osteryoung KW (2018) The Molecular Machinery of Chloroplast Division. Plant Physiol 176: 138-151
Dutta S, Cruz JA, Imran SM, Chen J, Kramer DM, Osteryoung KW (2017) Variations in chloroplast movement and chlorophyll fluorescence among chloroplast division mutants under light stress. J Exp Bot 68: 3541-3555
Honda SI, Hongladarom-Honda T, Kwanyuen P, Wildman SG (1971) Interpretations on chloroplast reproduction derived from correlations between cells and chloroplasts. Planta 97: 1-15
Jeffares DC, Tomiczek B, Sojo V, dos Reis M (2015) A beginners guide to estimating the non-synonymous to synonymous rate ratio of all protein-coding genes in a genome. Methods Mol Biol 1201: 65-90
Kadirjan-Kalbach DK, Turmo A, Wang J, Smith BC, Chen C, Porter KJ, Childs K, DellaPenna D, Osteryoung KW (2019) Allelic Variation in the Chloroplast Division Gene FtsZ2-2 Leads to Natural Variation in Chloroplast Size. Plant Physiol https://doi.org/10.1104/pp.19.00841
Keurentjes JJ, Bentsink L, Alonso-Blanco C, Hanhart CJ, Blankestijn-De Vries H, Effgen S, Vreugdenhil D, Koornneef M (2007) Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891-905
Miyagishima SY (2017) Chloroplast division: A handshake across membranes. Nat Plants 3: 17025
Schmitz AJ, Glynn JM, Olson BJ, Stokes KD, Osteryoung KW (2009) Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant 2: 1211-1222