De novo domestication of wild tomato using genome editing (Nature Biotech – $)
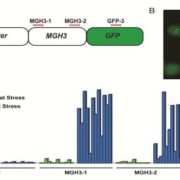
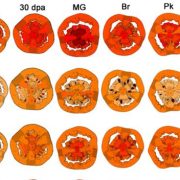
 Breeding of crops typically focuses on yield increase, and is accompanied with the loss the genetic diversity, reduced nutritional value as well as susceptibility to environmental stress. As many domestication traits exhibit simple Mendelian inheritance patterns, Zsögön et al. targeted known domestication loci in wild tomato, Solanum pimpinellifolium, using CRISPR/Cas9 genome editing technology. Successful targeting of SELF-PRUNING (SP), OVATE (O), FRUIT-WEIGHT2.2 (FW2.2) and LYCOPENE CYCLASE (CycB), resulted compact plant architecture but no change in fruit size. The second round of experiments targeted genes related to fruit shape, number and nutritional quality FW2.2, CycB, CLAVATA3 and MULTIFLORA, resulting in fourfold increase in fruit number and 200% increase of fruit weight. No editing of off-target loci was detected in any of the transformed plants. As the domestication traits were characterized for other crops, including maize, wheat and sorghum, targeted reverse genetic engineering has potential to boost the development of new crops. (Summary by Magdalena Julkowska) Nat. Biotech. 10.1038/nbt.4272
Breeding of crops typically focuses on yield increase, and is accompanied with the loss the genetic diversity, reduced nutritional value as well as susceptibility to environmental stress. As many domestication traits exhibit simple Mendelian inheritance patterns, Zsögön et al. targeted known domestication loci in wild tomato, Solanum pimpinellifolium, using CRISPR/Cas9 genome editing technology. Successful targeting of SELF-PRUNING (SP), OVATE (O), FRUIT-WEIGHT2.2 (FW2.2) and LYCOPENE CYCLASE (CycB), resulted compact plant architecture but no change in fruit size. The second round of experiments targeted genes related to fruit shape, number and nutritional quality FW2.2, CycB, CLAVATA3 and MULTIFLORA, resulting in fourfold increase in fruit number and 200% increase of fruit weight. No editing of off-target loci was detected in any of the transformed plants. As the domestication traits were characterized for other crops, including maize, wheat and sorghum, targeted reverse genetic engineering has potential to boost the development of new crops. (Summary by Magdalena Julkowska) Nat. Biotech. 10.1038/nbt.4272