Coevolving Chloroplast Proteins
By ES Forsythe, AM Williams, DB Sloan, Department of Biology, Colorado State University, Fort Collins, Colorado
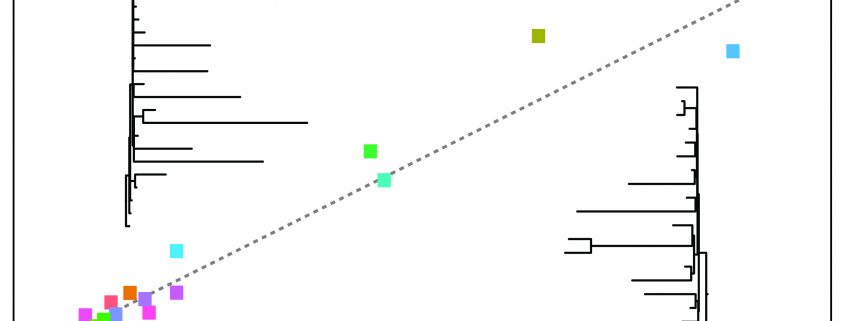
Background: Plant chloroplasts (plastids) have their own genome, which contains instructions for creating proteins that perform important work inside plastids. However, these proteins do not work alone; most of the proteins that function inside the plastids are encoded in the much larger nuclear genome and are imported into the plastids after being translated in the cytosol. Even though they come from different genomes, plastid- and nucleus-encoded proteins need to work together for plant survival. This type of interaction between proteins means that when one protein changes during evolution, natural selection also favors changes in their interacting proteins in order to maintain their coordination (i.e plastid-nuclear coevolution). As such, functionally related proteins are expected to exhibit correlated accelerations and decelerations in rates of amino acid sequence changes across species, which is known as evolutionary rate covariation (ERC). We used the expectation of ERC between interacting proteins to search the nuclear genomes of flowering plants to detect genes that are coevolving with the plastid genome and contribute to plastid function.
Question: Which nuclear genes coevolve with the plastid genome? Are there certain plastid processes that show especially prevalent plastid-nuclear coevolution?
Findings: We detected hundreds of nuclear genes that appear to coevolve with the plastid genome. Many of these genes encode known plastid-localized proteins but some have no identified plastid function, indicating that our analysis points to novel plastid functions for these genes. Genes involved in maintaining proper protein levels within the plastid (plastid proteostasis) appear to be especially prevalent, suggesting that changes to plastid proteostasis throughout the evolution of flowering plants may have driven plastid-nuclear coevolution. Surprisingly, this phenomenon even appears to extend to genes responsible for manufacturing proteins outside of plastids, meaning that even non-plastid-localized proteins contribute to plastid proteostasis and coevolve accordingly.
Next steps: Our work points to several genes playing newly discovered roles in plastids, representing high-priority candidates for experimental validation. Future genetic/molecular biology experiments will help us understand the specific functions of these proteins in the chloroplast.
Article: Evan S. Forsythe, Alissa M. Williams, and Daniel B. Sloan (2021). Genome-wide signatures of plastid-nuclear coevolution point to repeated perturbations of plastid proteostasis systems across angiosperms. Plant Cell, https://doi.org/10.1093/plcell/koab021