Co-evolution of the RNA editing factor PPR78 and its targets
Lesch et al. reveal that the mitochondrial RNA editing factor PPR78 of Physcomitrium patens is conserved among mosses despite the loss of its two known C-to-U editing sites due to the additional editing target ccmFNeU1465RC.
https://doi.org/10.1093/plcell/koad292
By Elena Lesch, Volker Knoop and Mareike Schallenberg-Rüdinger, IZMB – Institute for Cellular and Molecular Botany, Molecular Evolution Lab, University of Bonn
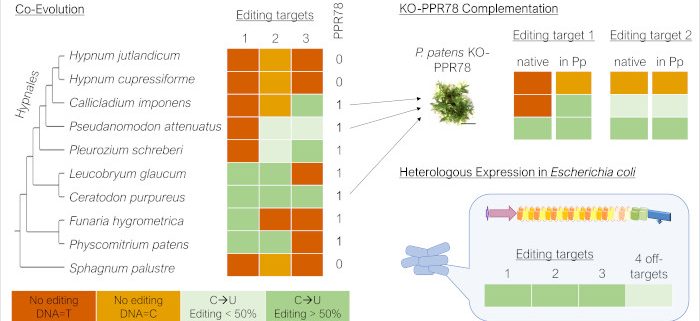
Background: Plant-type RNA editing refers to the site-specific deamination of cytidines to uridines in organellar transcripts. Pentatricopeptide repeat (PPR) proteins of the PLS subfamily are key players in this process. In mosses, PPR editing factors consist of an RNA binding PLS-type PPR array and a C-terminal DYW-type cytidine deaminase domain. PPR78 is one of nine organellar RNA editing factors in the model moss Physcomitrium patens. It edits two mitochondrial targets to different extents: the cox1eU755SL site is edited efficiently, whereas the rps14eU137SL site is edited partially. Establishing the cox1 site is also crucial in other mosses, whereas editing of rps14eU137SL is highly variable, ranging from non-existent to very efficient.
Question: We aimed to unravel the co-evolution of PPR78 and its two target sites in mosses. What causes the variability in rps14 editing efficiency? What happens to PPR78 if its two target sites are lost due to genomic C-T conversions creating a pre-edited state?
Findings: An early species sampling was widely extended, focusing on the Hypnales (feather mosses). In this moss order, the cox1 site is pre-edited, making editing obsolete, and the rps14 site is edited inefficiently or not at all while PPR78 orthologues remain conserved in most species. A complementation study of a Physcomitrium knockout mutant with PPR78 orthologues from different mosses revealed that variable rps14 editing is defined by the proteins themselves, specifically their PPR arrays. We successfully expressed functional PPR78 in Escherichia coli. Off-target profiling helped predict a third editing target site of PPR78 in Hypnales: ccmFNeU1465RC. We functionally assigned PPR78 to the ccmFN site in bacterio, providing evidence that PPR78 is evolutionarily retained in Hypnales to serve this additional editing target.
Next steps: In future, assignments of further editing sites to PPR78 may be explored. The region responsible for variable rps14eU137SL editing capacities of PPR78 orthologues may be narrowed down to certain PPR repeats. In addition, ccmFNeU1465RC editing functionalities of PPR78 orthologues may be tested by knockout studies in another moss or heterologous expression.
Reference:
Elena Lesch, Maike Simone Stempel, Vanessa Dressnandt, Bastian Oldenkott, Volker Knoop, and Mareike Schallenberg-Rüdinger. (2023). Conservation of the moss RNA editing factor PPR78 despite the loss of its known C-to-U editing sites is explained by a hidden extra target. https://doi.org/10.1093/plcell/koad292