Chimeric Activators and Repressors Define HY5 Activity
Early during seedling development and throughout their lives, plants sense and adapt to their constantly changing environment. Germinated seedlings perceive light which triggers a major developmental switch ultimately leading to photoautotrophic growth. This complex process, referred to broadly as photomorphogenesis, involves the perception of light through photoreceptors followed by a major reprogramming of the transcriptome (Chen et al., 2004). A complex network of transcription factors tightly controls these light-induced transcriptional programs. One key transcription factor is ELONGATED HYPOCOTYL 5 (HY5) and its absence results in plants not being able to respond properly to light (Koornneef et al., 1980). HY5 has been implicated in many biological processes by regulating the expression of thousands of genes (Gangappa and Botto, 2016). However to date, a consensus model for HY5-mediated transcriptional activation and/or repression is missing.
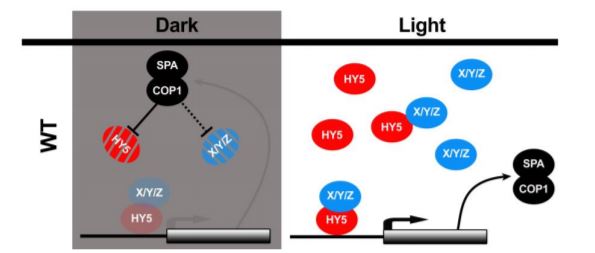
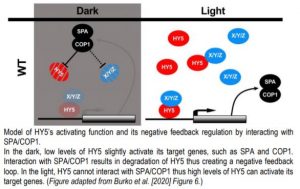 In this issue of The Plant Cell, Burko et al. (2020) investigated in detail how HY5 regulates transcription both in the dark and in the light. To do so, they leveraged transcriptionally activating or repressing HY5 chimeras with a stringent filtering strategy to define high confidence HY5 direct targets. The authors found that HY5 primarily works as a transcriptional activator that requires co-factors for activation. Furthermore, they showed that HY5 directly promotes the expression of SPA/COP1. In the dark, interaction with SPA/COP1 leads to the degradation of HY5 and its co-factors, thus creating a negative feedback loop that maintains HY5-mediated transcriptional activation at a low level (Figure).
In this issue of The Plant Cell, Burko et al. (2020) investigated in detail how HY5 regulates transcription both in the dark and in the light. To do so, they leveraged transcriptionally activating or repressing HY5 chimeras with a stringent filtering strategy to define high confidence HY5 direct targets. The authors found that HY5 primarily works as a transcriptional activator that requires co-factors for activation. Furthermore, they showed that HY5 directly promotes the expression of SPA/COP1. In the dark, interaction with SPA/COP1 leads to the degradation of HY5 and its co-factors, thus creating a negative feedback loop that maintains HY5-mediated transcriptional activation at a low level (Figure).
Several criteria were applied to establish that HY5 functions primarily as a transcriptional activator. First, HY5 binding sites were obtained by chromatin immunoprecipitation followed by sequencing (ChIP-seq). Next, the transcriptomes of plants expressing HY5-VP16 (chimeric activator) and HY5-SRDX (chimeric repressor) were profiled and compared to hy5 loss-of-function and HY5-overexpressing plants. Genes up- or -down-regulated in these four genetic backgrounds were cross-referenced to define 297 high-confidence HY5-regulated genes. Most of these genes were less expressed in the hy5 mutant indicating that HY5 primarily functions as a transcriptional activator. Finally, ChIP-seq measurements also showed that most of these 297 genes were bound by HY5, thus indicating that these genes are direct HY5 targets.
In darkness, HY5 is degraded by direct interaction with SPA/COP1 (Ang et al., 1998). During photomorphogenesis and deetiolation, COP1 is inactivated via the action of photoreceptors, leading to accumulation of HY5 that stimulates SPA and COP1 expression. Increased SPA and COP1 production then mitigates hyperaccumulation of HY5. To dissect this feedback loop, Burko and colleagues over-expressed truncated versions of HY5 and HY5-VP16 that are not recognized or degraded by SPA/COP1 (Figure). These results support the conclusion that HY5 requires SPA/COP1-targeted co-factors for transcriptional activation. This was further supported by the observation that, in contrast to HY5-VP16, HY5 alone was not able to activate a luciferase reporter in Drosophila S2 cells.
To expand the scope of their findings, the authors showed that chimeric HY5 activators and repressors can be used to alter tomato growth and development. This proof-of-principle experiment suggests that modulating HY5 levels could be an effective approach to control plant growth and improve crop productivity.
Sylvain Bischof
Department of Plant and Microbial Biology
University of Zürich, Switzerland
ORCHID ID: 0000-0003-2910-5132
REFERENCES
Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213-222.
Burko, Y., Seluzicki. A., Zander. M., Pedmale. U.V., Ecker. J.R., Chory. J. (2020). Chimeric Activators and Repressors Define HY5 Activity and Reveal a Light-Regulated Feedback Mechanism. Plant Cell. https://doi.org/10.1105/tpc.19.00772.
Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu Rev Genet 38: 87-117.
Gangappa, S.N., and Botto, J.F. (2016). The Multifaceted Roles of HY5 in Plant Growth and Development. Mol Plant 9: 1353-1365.
Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic Control of Light-inhibited Hypocotyl Elongation in Arabidopsis thaliana (L.) Heynh. Zeitschrift für Pflanzenphysiologie 100: 147-160.