Breaking the Mold: Transcriptome Analysis of Quantitative Disease Resistance to a Fungal Pathogen
Sucher et al. reveal that regulatory divergence of conserved genes makes a major contribution during the response of Pentapetalae plants to a fungal pathogen. Plant Cell. https://doi.org/10.1105/tpc.19.00806
Background:
Necrotrophic plant pathogens actively kill host cells to cause disease. This is notably the case for the fungus Sclerotinia sclerotiorum, causal agent of white and stem mold diseases on a broad range of dicot plants. Plants have developed specific resistance mechanisms to mitigate infections by necrotrophs, such as quantitative disease resistance (QDR). Studies on QDR so far have shown that this form of immunity is frequent in nature, often efficient against multiple pathogen genotypes (broad spectrum), and relatively stable over time (durable). QDR probably involves numerous genes with very diverse molecular functions, but the repertoire of genes contributing to QDR remains largely unknown.
Question:
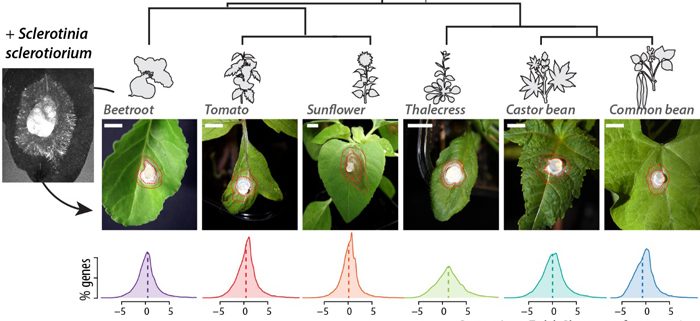
- sclerotiorum causes disease on plant species from very diverse botanical families in the Pentapetalae group of the dicots, such as common bean, castor bean, thalecress, tomato, sunflower and beetroot. How diverse are transcriptional responses to S. sclerotiorum in these plants? Are conserved genes and lineage-specific genes strongly regulated during infection?
Findings:
By studying the global transcriptome response of six plant species from diverse botanical families to the same strain of S. sclerotiorum, we showed that a large proportion of differentially express genes were conserved across species. However, despite being present in all species, “core” genes exhibited contrasting expression patterns during infection in each plant species.
These results suggested that some genes involved in QDR are broadly conserved stress-responsive genes that underwent changes in their transcriptional response to infection during evolution. We highlight one family of ABC transporter genes, conserved and induced by Sclerotinia infection in all plant species. Using Arabidopsis thaliana mutants, we show that ABCG40, the most induced member of this family, plays a role in QDR.
Next steps:
We identified the repertoire of genes conserved and responding similarly to fungal infection across botanical families, as a resource to mine for quantitative resistance genes active in multiple plant species. We wish now to investigate the molecular and evolutionary mechanisms underlying interspecific expression variation in QDR genes.
Justine Sucher, Malick Mbengue, Axel Dresen, Marielle Barascud, Marie Didelon, Adelin Barbacci, Sylvain Raffaele (2020) Phylotranscriptomics of the Pentapetalae Reveals Frequent Regulatory Variation in Plant Local Responses to the Fungal Pathogen Sclerotinia sclerotiorum. https://doi.org/10.1105/tpc.19.00806