A single-cell analysis of the Arabidopsis vegetative shoot apex (Devel. Cell)
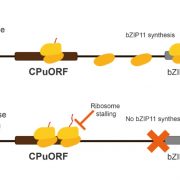
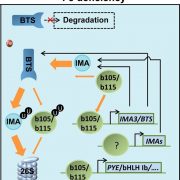
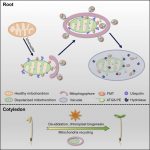
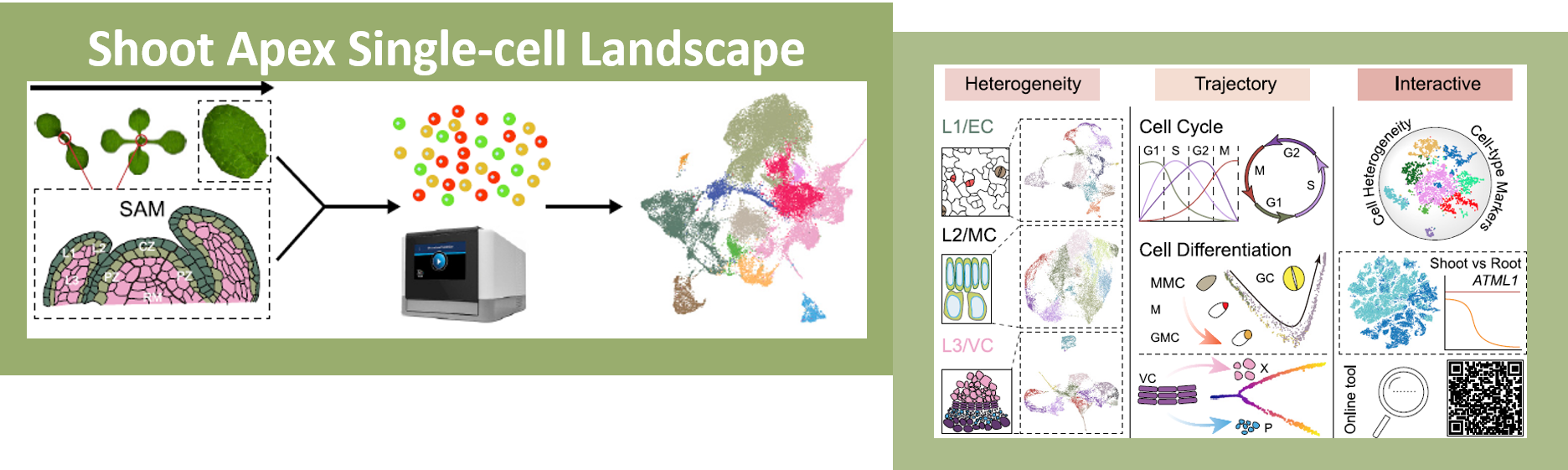 One of the most fascinating challenges in developmental biology is to understand how a few stem cells hidden in a microscopic region of the shoot apex give origin to all aerial parts of the plant. Technological advances such as single-cell RNA-sequencing (scRNA-seq) are enabling the comprehensive analysis of heterogeneous cell populations with unprecedent resolution. Zhang and coworkers previously employed scRNA-seq to determine the transcriptome profiles of the root apex in Arabidopsis. Here, they investigate stem cell functioning and cell fate determination of the shoot apex, and identified 23 cell clusters visualized into 7 broad populations. Transcriptome analysis of the shoot apex revealed accumulation of leaf polarity genes in a group of meristematic cells, indicating that these transit-amplification cells (i.e., in transition from undifferentiated to differentiated conditions) displayed a continuum of expression states during leaf primordia initiation. The authors also reconstructed developmental trajectories of different tissues within the shoot apex by combining spatial distribution and temporal ordering of individual cells, and arranged clusters in cyclic patterns depending on the transcript levels of core cell-cycle genes. Sequencing the transcriptomes of individual cells represents a revolutionary approach to decipher plant development and to accelerate the discovery of novel genes, with the resulting Root and Shoot Cell Atlases constituting valuable resources for the plant science community. You can search for your favorite genes at http://wanglab.sippe.ac.cn/shootatlas/. (Summary and image adaptation by Michela Osnato @michela_osnato) Dev. Cell. 10.1016/j.devcel.2021.02.021
One of the most fascinating challenges in developmental biology is to understand how a few stem cells hidden in a microscopic region of the shoot apex give origin to all aerial parts of the plant. Technological advances such as single-cell RNA-sequencing (scRNA-seq) are enabling the comprehensive analysis of heterogeneous cell populations with unprecedent resolution. Zhang and coworkers previously employed scRNA-seq to determine the transcriptome profiles of the root apex in Arabidopsis. Here, they investigate stem cell functioning and cell fate determination of the shoot apex, and identified 23 cell clusters visualized into 7 broad populations. Transcriptome analysis of the shoot apex revealed accumulation of leaf polarity genes in a group of meristematic cells, indicating that these transit-amplification cells (i.e., in transition from undifferentiated to differentiated conditions) displayed a continuum of expression states during leaf primordia initiation. The authors also reconstructed developmental trajectories of different tissues within the shoot apex by combining spatial distribution and temporal ordering of individual cells, and arranged clusters in cyclic patterns depending on the transcript levels of core cell-cycle genes. Sequencing the transcriptomes of individual cells represents a revolutionary approach to decipher plant development and to accelerate the discovery of novel genes, with the resulting Root and Shoot Cell Atlases constituting valuable resources for the plant science community. You can search for your favorite genes at http://wanglab.sippe.ac.cn/shootatlas/. (Summary and image adaptation by Michela Osnato @michela_osnato) Dev. Cell. 10.1016/j.devcel.2021.02.021