What We’re Reading: February 24
Insight: Why we need more non-seed plant models
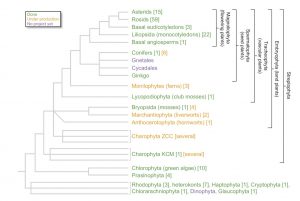 There is much to be learned from comparing plant genomes, but as Rensing writes, currently available genomic data are skewed heavily towards angiosperms. He argues that a richer understanding of plant evolution depends upon gaining insights into the non-seed plants, including ferns, mosses and liverworts, but particularly the different lineages of charophyte algae, some of which share a common ancestor with land plants. In addition to sequenced genomes, genetic tools such as genome editing and complementation assays will be needed to infer gene functional relationships. New Phytol. 10.1111/nph.14464 Tags: Botany, Evolution, Genomics
There is much to be learned from comparing plant genomes, but as Rensing writes, currently available genomic data are skewed heavily towards angiosperms. He argues that a richer understanding of plant evolution depends upon gaining insights into the non-seed plants, including ferns, mosses and liverworts, but particularly the different lineages of charophyte algae, some of which share a common ancestor with land plants. In addition to sequenced genomes, genetic tools such as genome editing and complementation assays will be needed to infer gene functional relationships. New Phytol. 10.1111/nph.14464 Tags: Botany, Evolution, Genomics
Perspective: Periodic lateral root priming: What makes it tick
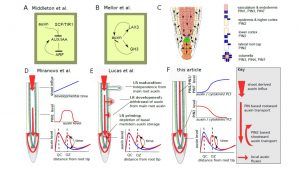 The very earliest step in lateral root formation is “priming”, a still-elusive process that establishes a subset of cells as competent to form lateral roots. In this Perspective, ten Tusscher and Laskowski review evidence for priming, which involves oscillations of auxin level or responsiveness and oscillations of gene expression. The authors describe several models for the origin of these oscillations, including cell-autonomous clocks, Turing-type patterning, and tissue-level alterations in auxin level. The authors conclude by presenting a new hypothesis, “in which self-organized fluctuations in auxin level both result in and arise from changes in auxin transporters.” Plant Cell 10.1105/tpc.16.00638 Tags: Computational Biology, Development, Signals and Responses
The very earliest step in lateral root formation is “priming”, a still-elusive process that establishes a subset of cells as competent to form lateral roots. In this Perspective, ten Tusscher and Laskowski review evidence for priming, which involves oscillations of auxin level or responsiveness and oscillations of gene expression. The authors describe several models for the origin of these oscillations, including cell-autonomous clocks, Turing-type patterning, and tissue-level alterations in auxin level. The authors conclude by presenting a new hypothesis, “in which self-organized fluctuations in auxin level both result in and arise from changes in auxin transporters.” Plant Cell 10.1105/tpc.16.00638 Tags: Computational Biology, Development, Signals and Responses
Reviews: Challenges in bacterial molecular plant pathology
 Molecular Plant Pathology has released a new series of free reviews on “Challenges in Bacterial Molecular Plant Pathology.” Topics so far:
Molecular Plant Pathology has released a new series of free reviews on “Challenges in Bacterial Molecular Plant Pathology.” Topics so far:
- Morris et al. Frontiers for research on the ecology of plant-pathogenic bacteria: fundamentals for sustainability 1111/mpp.12508
- Pfeilmeier et al. Bacterial pathogenesis of plants: future challenges from a microbial perspective 1111/mpp.12427
- Sundin et al. Bacterial disease management: challenges, experience, innovation and future prospects 1111/mpp.12436
- Baltrus et al. Evolution, genomics and epidemiology of Pseudomonas syringae 1111/mpp.12506
- Profiling the extended phenotype of plant pathogens 10.1111/mpp.12530
Tags: Applied Plant Biology, Biotic Interactions, Education and Outreach, Evolution
Meeting Review: Auxin 2016 ($)
 Vernoux and Robert discuss research presented at the Auxin 2016 meeting, held in October 2016 on the island of Hainan, China. The result is a nice review of very current research, spanning auxin metabolism and signaling, the role of auxin in development, interactions between auxin and other signals, and modeling and synthetic biology approaches to understand auxin function. Development 10.1242/dev.144790 Tags: Computational Biology, Development, Gene Regulation, Signals and Responses,
Vernoux and Robert discuss research presented at the Auxin 2016 meeting, held in October 2016 on the island of Hainan, China. The result is a nice review of very current research, spanning auxin metabolism and signaling, the role of auxin in development, interactions between auxin and other signals, and modeling and synthetic biology approaches to understand auxin function. Development 10.1242/dev.144790 Tags: Computational Biology, Development, Gene Regulation, Signals and Responses,
Review: Stomatal development in time: the past and the future ($)
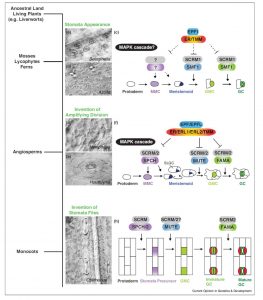 Stomata, epidermal pores for gas exchange, first appeared about 400 million years ago. Since then, there has been functional and structural diversification. Qu et al. synthesize the developmental genetics underpinning diverse stomata, spanning from bryophytes through monocots and the astomatous (without stomata) marine angiosperms, as revealed by seagrass. Curr. Opin. Genet. Devel. 10.1016/j.gde.2017.02.001 Tags: Botany, Cell Biology, Development, Evolution, Signals and Responses
Stomata, epidermal pores for gas exchange, first appeared about 400 million years ago. Since then, there has been functional and structural diversification. Qu et al. synthesize the developmental genetics underpinning diverse stomata, spanning from bryophytes through monocots and the astomatous (without stomata) marine angiosperms, as revealed by seagrass. Curr. Opin. Genet. Devel. 10.1016/j.gde.2017.02.001 Tags: Botany, Cell Biology, Development, Evolution, Signals and Responses
Review: Nitrogen sensing in plants ($)
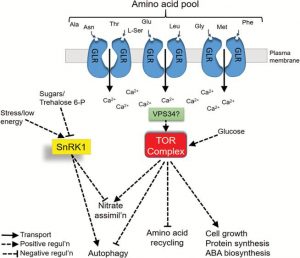 Like other organisms, plants have developed mechanisms to sense and respond to changes in the availability of nutrients. Nitrogen (N), being very essential for the growth and development of plants, must also be strongly monitored by plants. N sensing and signaling in plants are highly researched topics, but uncertainty remains in the nature of N-sensing mechanisms for controlling N uptake and plant development. In this review, Gent and Forde discuss potential N sensing molecules and pathway including glutamine, TOR complex and GLR family of glutamate-like receptors (GLRs). (Summary by Mather Khan) J. Exp. Bot. 10.1093/jxb/erx013 Tags: Biochemistry, Gene Regulation, Signals and Responses, Transport
Like other organisms, plants have developed mechanisms to sense and respond to changes in the availability of nutrients. Nitrogen (N), being very essential for the growth and development of plants, must also be strongly monitored by plants. N sensing and signaling in plants are highly researched topics, but uncertainty remains in the nature of N-sensing mechanisms for controlling N uptake and plant development. In this review, Gent and Forde discuss potential N sensing molecules and pathway including glutamine, TOR complex and GLR family of glutamate-like receptors (GLRs). (Summary by Mather Khan) J. Exp. Bot. 10.1093/jxb/erx013 Tags: Biochemistry, Gene Regulation, Signals and Responses, Transport
Review: Winter and summer dormancy: similar adaptive strategies?
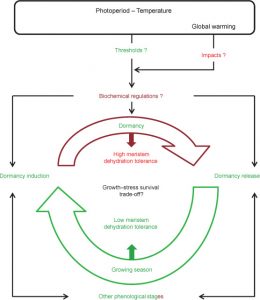 Dormancy (growth arrest) is a state by which seeds and plants can survive harsh conditions. Seasonal dormancy is a strategy to survive seasonally unfavorable conditions. Plants can display winter and summer dormancy. Although woody species are the main study systems for winter dormancy, herbaceous species show us that both kinds of dormancy share some similarities, as well as some differences. Gillespie and Volaire describe what is known about the mechanisms of winter and summer dormancy, as well as the biochemical mechanisms involved. With projections of climate change, a deeper understanding seems increasingly relevant. Will plants start expressing more summer dormancy traits to adapt? (Summary by Gabe Auge) Ann. Bot. 10.1093/aob/mcw264 Tags: Abiotic Interactions, Botany, Biochemistry, Ecophysiology, Metabolism, Signals and Responses
Dormancy (growth arrest) is a state by which seeds and plants can survive harsh conditions. Seasonal dormancy is a strategy to survive seasonally unfavorable conditions. Plants can display winter and summer dormancy. Although woody species are the main study systems for winter dormancy, herbaceous species show us that both kinds of dormancy share some similarities, as well as some differences. Gillespie and Volaire describe what is known about the mechanisms of winter and summer dormancy, as well as the biochemical mechanisms involved. With projections of climate change, a deeper understanding seems increasingly relevant. Will plants start expressing more summer dormancy traits to adapt? (Summary by Gabe Auge) Ann. Bot. 10.1093/aob/mcw264 Tags: Abiotic Interactions, Botany, Biochemistry, Ecophysiology, Metabolism, Signals and Responses
A novel chemical inhibitor of ABA signaling targets all ABA receptors
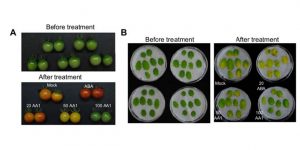 Chemical genetics is an approach that enables small molecules with specific effects on phenotypes to be identified. Through a screen for small molecules that would reverse the inhibitory effect of ABA on seed germination, Ye et al. identified AA1 (ABA Antagonist 1). AA1 interferes with the interaction between all of the ABA PYR-PYL receptors and their associated PP2C phosphatases. AA1 is a broad-spectrum ABA antagonist that inhibits numerous ABA responses including gene expression, leaf senescence, fruit ripening and stomatal closure, providing a new tool for research and agriculture. Plant Physiol. 10.1104/pp.16.01862 Tags: Biochemistry, Gene Regulation, Genetics, Physiology
Chemical genetics is an approach that enables small molecules with specific effects on phenotypes to be identified. Through a screen for small molecules that would reverse the inhibitory effect of ABA on seed germination, Ye et al. identified AA1 (ABA Antagonist 1). AA1 interferes with the interaction between all of the ABA PYR-PYL receptors and their associated PP2C phosphatases. AA1 is a broad-spectrum ABA antagonist that inhibits numerous ABA responses including gene expression, leaf senescence, fruit ripening and stomatal closure, providing a new tool for research and agriculture. Plant Physiol. 10.1104/pp.16.01862 Tags: Biochemistry, Gene Regulation, Genetics, Physiology
Molecular signals for regeneration ($)
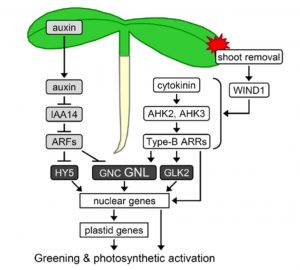 Every gardener knows that pulling off a shoot is not sufficient to kill a plant. Plants are able to reprogram cells in order to regenerate missing tissues. Pulling off a shoot removes photosynthetic tissues, but the root responds by activating chloroplasts to take over this role. Kobayashi et al. (Plant Physiol. 10.1104/pp.16.01368) examine the signals involved in chloroplast activation in roots following shoot removal; wound responses, cytokinin increase and auxin depletion all contribute to this reprogramming. To a large extent, regeneration involves activating differentiation pathways. Previously, nuclear accumulation of the transcription factor IYO was shown to trigger cell differentiation. Muñoz et al. (Plant Cell 10.1105/tpc.16.00791) identified a nuclear import protein RIMA involved in nuclear accumulation of IYO; this nuclear import triggers auxin-irreversible cell differentiation in Arabidopsis. Furthemore, that fact that IYO is excluded from the nucleus in xylem-pole pericycle cells supports their proposed role as serving as undifferentiated stem-cell reservoirs. Tags: Bioenergetics, Development, Signals and Responses
Every gardener knows that pulling off a shoot is not sufficient to kill a plant. Plants are able to reprogram cells in order to regenerate missing tissues. Pulling off a shoot removes photosynthetic tissues, but the root responds by activating chloroplasts to take over this role. Kobayashi et al. (Plant Physiol. 10.1104/pp.16.01368) examine the signals involved in chloroplast activation in roots following shoot removal; wound responses, cytokinin increase and auxin depletion all contribute to this reprogramming. To a large extent, regeneration involves activating differentiation pathways. Previously, nuclear accumulation of the transcription factor IYO was shown to trigger cell differentiation. Muñoz et al. (Plant Cell 10.1105/tpc.16.00791) identified a nuclear import protein RIMA involved in nuclear accumulation of IYO; this nuclear import triggers auxin-irreversible cell differentiation in Arabidopsis. Furthemore, that fact that IYO is excluded from the nucleus in xylem-pole pericycle cells supports their proposed role as serving as undifferentiated stem-cell reservoirs. Tags: Bioenergetics, Development, Signals and Responses
Allelic diversity underlying flowering-time adaptation in maize landraces ($)
 Landraces are native varieties that have been selected for adaptation to their native environment, and as such they provide a wealth of largely unexplored genetic potential. Romero Navarro et al. used a new approach called F-one association mapping (FOAM) in combination with genome-wide association strategy (GWAS) to query an initial population of nearly 5000 diverse individuals from 35 countries. Flowering time is a multi-genic trait crucial for local adaptation and particularly responsive to latitude and altitude. Through this study, the authors identified nearly 1500 genes associated with altitude, and more than 1000 associated with flowering, demonstrating that this approach is a powerful tool with which to unlock the information stored in landrace genomes. Nature Genetics 10.1038/ng.3784 Tags: Ecophysiology, Genetics, Genomics
Landraces are native varieties that have been selected for adaptation to their native environment, and as such they provide a wealth of largely unexplored genetic potential. Romero Navarro et al. used a new approach called F-one association mapping (FOAM) in combination with genome-wide association strategy (GWAS) to query an initial population of nearly 5000 diverse individuals from 35 countries. Flowering time is a multi-genic trait crucial for local adaptation and particularly responsive to latitude and altitude. Through this study, the authors identified nearly 1500 genes associated with altitude, and more than 1000 associated with flowering, demonstrating that this approach is a powerful tool with which to unlock the information stored in landrace genomes. Nature Genetics 10.1038/ng.3784 Tags: Ecophysiology, Genetics, Genomics
Durable resistance gene Xa4 encodes a cell wall-associated kinase
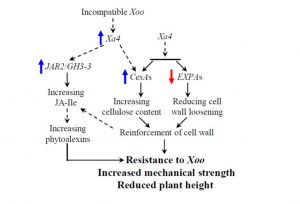 Xa4 is a durable rice disease resistance gene that confers resistance against Xanthomonas oryzae pv. oryzae (Xoo), causal agent of bacterial blight. Hu et al. show that Xa4 encodes a wall-associated kinase (WAK) that promotes cellulose synthesis and suppresses wall loosening, thereby strengthening the cell wall. Suppression of disease is due to Xa4’s effects on cell wall integrity. Furthermore, Xa4 is associated with shorter stature, a beneficial agronomic trait that helps prevent lodging. As the authors say, “The simultaneous improvement of multiple agronomic traits conferred by Xa4 may account for its idespread and lasting utilization in rice breeding programmes globally.” Nature Plants 10.1038/nplants.2017.9 Tags: Biotic Interactions, Cell Biology, Growth Regulation
Xa4 is a durable rice disease resistance gene that confers resistance against Xanthomonas oryzae pv. oryzae (Xoo), causal agent of bacterial blight. Hu et al. show that Xa4 encodes a wall-associated kinase (WAK) that promotes cellulose synthesis and suppresses wall loosening, thereby strengthening the cell wall. Suppression of disease is due to Xa4’s effects on cell wall integrity. Furthermore, Xa4 is associated with shorter stature, a beneficial agronomic trait that helps prevent lodging. As the authors say, “The simultaneous improvement of multiple agronomic traits conferred by Xa4 may account for its idespread and lasting utilization in rice breeding programmes globally.” Nature Plants 10.1038/nplants.2017.9 Tags: Biotic Interactions, Cell Biology, Growth Regulation
1135 Arabidopsis genomes reveal global pattern of polymorphism
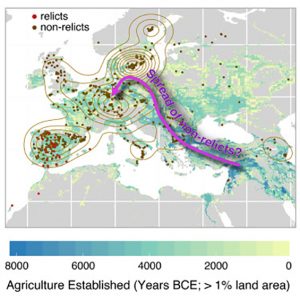 There are many accessions of Arabidopsis thaliana beyond the ecotypes predominantly used in research laboratories. In this article, The 1001 Genomes Consortium describe a resource based on whole-genome sequencing of 1,135 A. thaliana genomes from Europe, North Africa, and North America. This data set provides high-quality genomic data suitable for GWAS and population genetics studies. Resources from this project are available through web-based tools at http://1001genomes.org/tools/, which can be used to answer questions on genetic diversity and environmental adaptations. For example, SNPs correlated with climate factors were identified related to drought response and fungal defense. Furthermore, ancient expansion of glaciers has shaped A. thaliana populations, and most extant A. thaliana populations can be traced to a single population expansion event when species from North Africa colonized Europe and the British Isles. A limited number of relict A. thaliana populations exist in geographic areas, such as the Iberian Peninsula, that have experienced minimal changes in climate since the last glaciers occupied these areas. A major conclusion from analysis of these 1,135 genomes is A. thaliana contains DNA from multiple ancestors, but a single historical population expansion has favored descendants from one geographic area, which is still unknown. (Summary by Daniel Czerny) Cell 10.1016/j.cell.2016.05.063 Tags: Botany, Ecophysiology, Evolution, Genomics
There are many accessions of Arabidopsis thaliana beyond the ecotypes predominantly used in research laboratories. In this article, The 1001 Genomes Consortium describe a resource based on whole-genome sequencing of 1,135 A. thaliana genomes from Europe, North Africa, and North America. This data set provides high-quality genomic data suitable for GWAS and population genetics studies. Resources from this project are available through web-based tools at http://1001genomes.org/tools/, which can be used to answer questions on genetic diversity and environmental adaptations. For example, SNPs correlated with climate factors were identified related to drought response and fungal defense. Furthermore, ancient expansion of glaciers has shaped A. thaliana populations, and most extant A. thaliana populations can be traced to a single population expansion event when species from North Africa colonized Europe and the British Isles. A limited number of relict A. thaliana populations exist in geographic areas, such as the Iberian Peninsula, that have experienced minimal changes in climate since the last glaciers occupied these areas. A major conclusion from analysis of these 1,135 genomes is A. thaliana contains DNA from multiple ancestors, but a single historical population expansion has favored descendants from one geographic area, which is still unknown. (Summary by Daniel Czerny) Cell 10.1016/j.cell.2016.05.063 Tags: Botany, Ecophysiology, Evolution, Genomics



Leave a Reply
Want to join the discussion?Feel free to contribute!