The final (algal) frontier: Chlamydomonas enters the age of single cell RNA-seq
By Patrice A. Salomé, Department of Chemistry and Biochemistry, UCLA*
Background: The sequenced genome of the unicellular photosynthetic alga Chlamydomonas reinhardtii opened the doors for a multitude of transcriptome deep sequencing (RNA-seq) experiments, thanks to the ease with which this alga can be grown in precise growth media. However, all RNA-seq data to date have been generated from bulk cultures, to collect sufficient material for RNA extraction. In addition, most cultures are grown and collected in constant light, with the untested hypothesis that these cultures are globally asynchronous.
Question: Can we extract total RNA from single C. reinhardtii cells that is suitable for sequencing by single cell (sc)RNA-seq? Can we make biological sense of the transcript profiles? Finally, does the presence of the cell wall in some Chlamydomonas laboratory strains, and presumably in natural algal communities, interfere with RNA extraction or analysis? We chose to profile two strains (one with, and one without a cell wall) grown in three conditions: full supply of iron and nitrogen; iron limitation alone; nitrogen limitation alone.
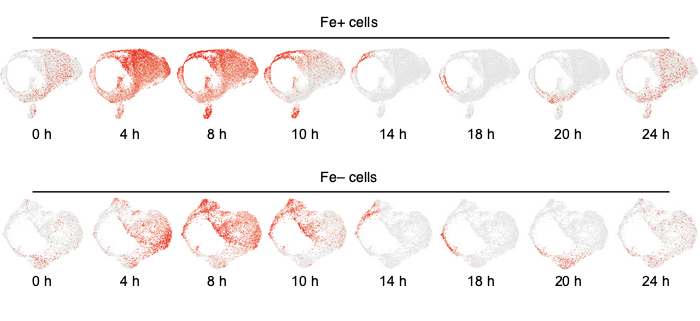
Findings: We sequenced close to 60,000 individual Chlamydomonas cells across all genotypes and growth conditions. Clustering of the high dimension transcriptome data we obtained clearly separated cells based on the treatment they were exposed to. Surprisingly, we noticed that replete cells for iron and nitrogen were strongly rhythmic and largely shared the same diurnal phase, although the last dark-to light transition they experienced had been 5 days prior. Furthermore, although iron-limited cells were also largely rhythmic individually, they collective diurnal phase was shifted later, results that would indicate the potential conservation of iron circadian responses from algae to land plants. Finally, the cell wall did not prevent the effective isolation or sequencing of RNA.
Next steps: Since most bulk RNA-seq data are generated from cultures grown in constant light, and we showed here that such cultures remain not only rhythmic at the individual cell level, as expected, but also largely synchronous, it will be important to determine how long such cultures can stay in sync in the absence of obvious entraining cues. The application of scRNA-seq to algal specimens collecting in the wild will also be a priority.
Reference:
Feiyang Ma, Patrice A. Salomé, Sabeeha S. Merchant and Matteo Pellegrini (2021). Single-Cell RNA Sequencing of Batch Chlamydomonas Cultures Reveals Heterogeneity in their Diurnal Cycle Phase. The Plant Cell, https://doi.org/10.1093/plcell/koab025
*Current affiliation: Science Editor, ASPB