Single-residue substitution in histone H3 leads to over-lignification in Arabidopsis
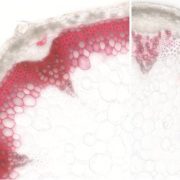
 Chromatin changes are at the core of plant development and adaptation, as they constitute a crucial layer of gene expression control. The effects of different chromatin marks have historically been studied through mutations to the protein complexes depositing and removing the marks, which results in strong developmental defects and pleiotropic effects. Fal et al. expressed a histone H3 variant (H3.3) with a mutation in lysine 27 (lysine to alanine, or K27A) that prevents its modification. Arabidopsis plants expressing H3.3K27A displayed strong phenotypes, some of them resembling polycomb repressive complex 2 mutants (shorter stems, twisted leaves and early flowering) and some new ones, such as a marked thickening of the lignified tissue. A combination of transcriptomics and metabolomics analyses revealed that almost 70% of phenol-related metabolites mis-accumulated in H3.3K27A were phenylpropanoids, and that over 20% of genes involved in phenylpropanoid biosynthesis were differentially expressed in H3.3K27A. As phenylpropanoids are core components of lignin, the misregulation of their synthesis is likely to be linked to the observed over-lignification. Extending this strategy to additional histone residues could give new insights into the redundant and combinatorial nature of chromatin modifications. (Summary by Laura Turchi @turchi_l) New Phytol. 10.1111/nph.18666
Chromatin changes are at the core of plant development and adaptation, as they constitute a crucial layer of gene expression control. The effects of different chromatin marks have historically been studied through mutations to the protein complexes depositing and removing the marks, which results in strong developmental defects and pleiotropic effects. Fal et al. expressed a histone H3 variant (H3.3) with a mutation in lysine 27 (lysine to alanine, or K27A) that prevents its modification. Arabidopsis plants expressing H3.3K27A displayed strong phenotypes, some of them resembling polycomb repressive complex 2 mutants (shorter stems, twisted leaves and early flowering) and some new ones, such as a marked thickening of the lignified tissue. A combination of transcriptomics and metabolomics analyses revealed that almost 70% of phenol-related metabolites mis-accumulated in H3.3K27A were phenylpropanoids, and that over 20% of genes involved in phenylpropanoid biosynthesis were differentially expressed in H3.3K27A. As phenylpropanoids are core components of lignin, the misregulation of their synthesis is likely to be linked to the observed over-lignification. Extending this strategy to additional histone residues could give new insights into the redundant and combinatorial nature of chromatin modifications. (Summary by Laura Turchi @turchi_l) New Phytol. 10.1111/nph.18666