Single cell transcriptomics after mild drought reveals two types of mesophyll responses
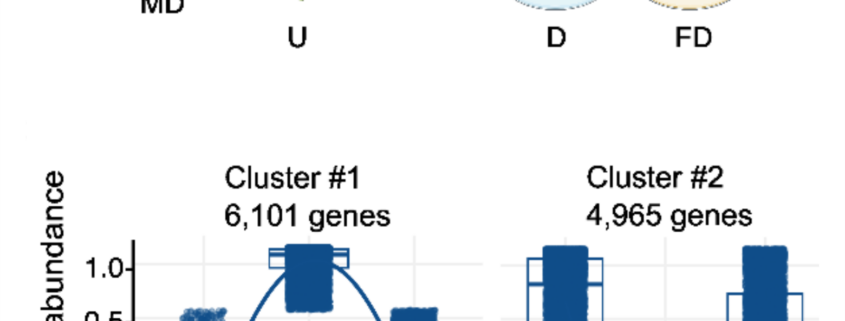
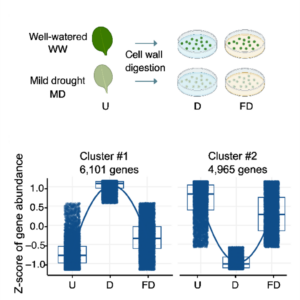 Single-cell transcriptomics offer unprecedented insights into how plants respond to their environment. Although this technique is powerful, it requires extensive processing of the tissue (cell wall digestion to form protoplasts) prior to sequencing, which can induce additional transcriptional changes. Here, Tenorio Berrío et al. incorporated into their protocol a block on transcription through applying actinomycin D (ActD) prior to cell isolation. Using this method, they investigated gene expression in young Arabidopsis leaves following mild drought. They compared these new results to those from a prior study they had conducted without the transcriptional block and found significant differences in gene expression. Interestingly, many known drought-response genes were absent from the samples that had been digested without the ActD, suggesting that the transcriptional response to cell wall degradation masked the transcriptional response to drought. They followed this up with whole-mount in situ hybridization studies of several genes. From the dataset obtained from the ActD treated sample, they identified two subpopulations of mesophyll cells as defined by their transcriptomes. One, located around the leaf margin and near the veins, is characterized by drought-responsive genes. A spatially distinct set, closer to the main photosynthetic cells, showed an upregulation of iron starvation-responsive genes; the authors discuss the potential origin of this response. The article illustrates the value of using ActD to arrest transcription prior to single cell isolation. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.08.30.61043
Single-cell transcriptomics offer unprecedented insights into how plants respond to their environment. Although this technique is powerful, it requires extensive processing of the tissue (cell wall digestion to form protoplasts) prior to sequencing, which can induce additional transcriptional changes. Here, Tenorio Berrío et al. incorporated into their protocol a block on transcription through applying actinomycin D (ActD) prior to cell isolation. Using this method, they investigated gene expression in young Arabidopsis leaves following mild drought. They compared these new results to those from a prior study they had conducted without the transcriptional block and found significant differences in gene expression. Interestingly, many known drought-response genes were absent from the samples that had been digested without the ActD, suggesting that the transcriptional response to cell wall degradation masked the transcriptional response to drought. They followed this up with whole-mount in situ hybridization studies of several genes. From the dataset obtained from the ActD treated sample, they identified two subpopulations of mesophyll cells as defined by their transcriptomes. One, located around the leaf margin and near the veins, is characterized by drought-responsive genes. A spatially distinct set, closer to the main photosynthetic cells, showed an upregulation of iron starvation-responsive genes; the authors discuss the potential origin of this response. The article illustrates the value of using ActD to arrest transcription prior to single cell isolation. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.08.30.61043