Review: Unlocking plant genetics with telomere-to-telomere genome assemblies
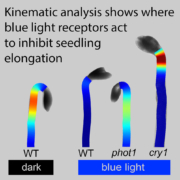
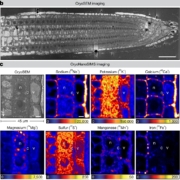
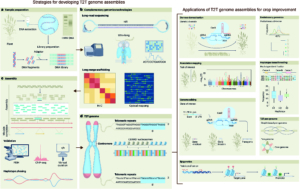 The era of complete telomere-to-telomere (T2T) plant genomes is finally here! Recent advances in long-read sequencing technologies have revolutionized genome assembly, giving rise to gapless T2T assemblies and offering unprecedented insights into genome organization and function. Why are T2T genomes so important? These assemblies provide a comprehensive, end-to-end view of a genome, covering all coding and non-coding regions. However, constructing T2T assemblies, especially for complex crop genomes with challenges like repeats, polyploidy, and heterozygosity, is a difficult task. This review by Garg et al. highlights such challenges and delves into the strategies and technological advancements necessary to overcome them. High-quality DNA extraction, improved sequencing accuracy, and the use of long-range scaffolding techniques like high-throughput chromosome conformation capture (Hi-C) and optical mapping are critical. Combining long-read sequencing and assembly algorithms, followed by manual curation, has resulted in complete T2T assemblies for crops such as rice and maize. The review also describes the current status of T2T plant genomes and their applications, including pangenomics, functional gene discovery, QTL cloning, and breeding strategies. Complete T2T assemblies promise to resolve genetic complexities in important traits like yield, disease resistance, and climate adaptation and open up new research possibilities, including developing new crop varieties for future needs. However, achieving these assemblies requires collaboration among geneticists, bioinformaticians, breeders, and other experts, along with continued investment in sequencing technologies and data integration. (Summary by Ileana Tossolini @IleanaDrt) Nature Genetics 10.1038/s41588-024-01830-7
The era of complete telomere-to-telomere (T2T) plant genomes is finally here! Recent advances in long-read sequencing technologies have revolutionized genome assembly, giving rise to gapless T2T assemblies and offering unprecedented insights into genome organization and function. Why are T2T genomes so important? These assemblies provide a comprehensive, end-to-end view of a genome, covering all coding and non-coding regions. However, constructing T2T assemblies, especially for complex crop genomes with challenges like repeats, polyploidy, and heterozygosity, is a difficult task. This review by Garg et al. highlights such challenges and delves into the strategies and technological advancements necessary to overcome them. High-quality DNA extraction, improved sequencing accuracy, and the use of long-range scaffolding techniques like high-throughput chromosome conformation capture (Hi-C) and optical mapping are critical. Combining long-read sequencing and assembly algorithms, followed by manual curation, has resulted in complete T2T assemblies for crops such as rice and maize. The review also describes the current status of T2T plant genomes and their applications, including pangenomics, functional gene discovery, QTL cloning, and breeding strategies. Complete T2T assemblies promise to resolve genetic complexities in important traits like yield, disease resistance, and climate adaptation and open up new research possibilities, including developing new crop varieties for future needs. However, achieving these assemblies requires collaboration among geneticists, bioinformaticians, breeders, and other experts, along with continued investment in sequencing technologies and data integration. (Summary by Ileana Tossolini @IleanaDrt) Nature Genetics 10.1038/s41588-024-01830-7