PCMD: an interactive library for comparative metabolomics studies
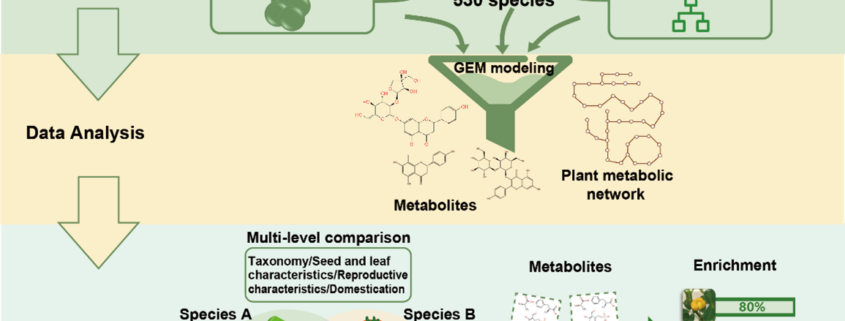
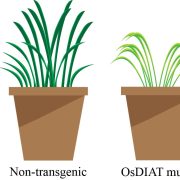
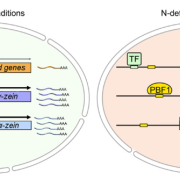
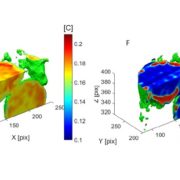
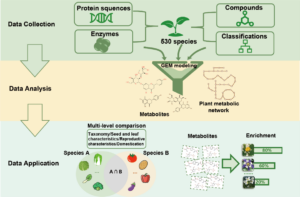 Albert Einstein once said, “The only thing that you absolutely have to know is the location of the library.” Libraries house vast troves of information for readers to explore, analyze, and use. With the exponential increase in data, libraries have also evolved into digital databases and online platforms. For example, large-scale omics studies yield tremendous amounts of data on genes, proteins, and metabolites that can help us understand plant development, provided they are accessible to researchers. In a recent issue of Plant Communications, Hu and colleagues introduced the Plant Comparative Metabolome Database (PCMD; https://yanglab.hzau.edu.cn/PCMD) for comparative metabolomics. Built on genome-based predictions of metabolites and associated metabolic pathways, along with supporting experimental data, PCMD provides metabolic profiles for 530 plant species. It offers unique features such as metabolite enrichment determination for cross-species comparative analysis, setting it apart from other databases. Each metabolite entry also includes data on associated proteins, metabolic reactions, and relevant literature, and links to databases like PubChem and MetaCyc. Future updates include tools for uploading experimental data and visualizing metabolic networks to deepen studies of gene-metabolite relationships. In conclusion, like a library, PCMD provides researchers interested in comparative metabolomics with a robust starting point for exploring metabolites, metabolic profiles, and the evolution of metabolic networks and can even support biotechnological research focused on plant-derived compounds for pharmaceutical or therapeutic applications. (Summary by Thomas Depaepe @thdpaepe) Plant Communications 10.1016/j.xplc.2024.101038.
Albert Einstein once said, “The only thing that you absolutely have to know is the location of the library.” Libraries house vast troves of information for readers to explore, analyze, and use. With the exponential increase in data, libraries have also evolved into digital databases and online platforms. For example, large-scale omics studies yield tremendous amounts of data on genes, proteins, and metabolites that can help us understand plant development, provided they are accessible to researchers. In a recent issue of Plant Communications, Hu and colleagues introduced the Plant Comparative Metabolome Database (PCMD; https://yanglab.hzau.edu.cn/PCMD) for comparative metabolomics. Built on genome-based predictions of metabolites and associated metabolic pathways, along with supporting experimental data, PCMD provides metabolic profiles for 530 plant species. It offers unique features such as metabolite enrichment determination for cross-species comparative analysis, setting it apart from other databases. Each metabolite entry also includes data on associated proteins, metabolic reactions, and relevant literature, and links to databases like PubChem and MetaCyc. Future updates include tools for uploading experimental data and visualizing metabolic networks to deepen studies of gene-metabolite relationships. In conclusion, like a library, PCMD provides researchers interested in comparative metabolomics with a robust starting point for exploring metabolites, metabolic profiles, and the evolution of metabolic networks and can even support biotechnological research focused on plant-derived compounds for pharmaceutical or therapeutic applications. (Summary by Thomas Depaepe @thdpaepe) Plant Communications 10.1016/j.xplc.2024.101038.