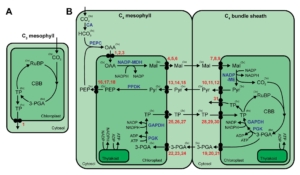
Double trouble: The Solanum pan-genome shows gene duplication complicates predictability
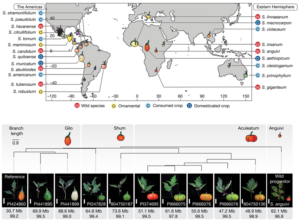 A pan-genome has been assembled for the Solanum genus, which contains many diverse and economically important crops including potato, tomato, and African eggplant. Genomes were assembled for 22 species, and genes were predicted based on previous reference genomes and from RNA sequences across multiple tissues. Approximately 60% of genes were the core genome (found in all species), and while synteny was mostly conserved across the species, large scale inversions and translocations were seen within subclades. While gene content was largely consistent between species, genome sizes ranged from 0.7-2.5 Gb mainly due to retrotransposon activity, but also from gene duplications. Over half a million duplications were found, both whole genome and single-gene, and paralogues disproportionally evolved to maintain normal gene dosage. This was greatly achieved through conservation of the cis-regulatory sequences compared to the coding portion of genes through evolution. Paralogues with similar expression patterns may be partially or fully redundant, compensatory, or pseudogenized, which may mean related genes in another species have different functions and effects on plant phenotypes. This complicates genetic engineering for crop improvement, as the effect on phenotype in one species may not reliably predict the outcome in others. However, with the continual establishment of lineage-specific pan-genomes, combined with advances in machine learning models, enhancement in the prediction accuracy is anticipated, paving the way for more precise and efficient crop improvement strategies. (Summary by Ciara O’Brien @ciara.obrien.bsky.social) Nature 10.1038/s41586-025-08619-6
A pan-genome has been assembled for the Solanum genus, which contains many diverse and economically important crops including potato, tomato, and African eggplant. Genomes were assembled for 22 species, and genes were predicted based on previous reference genomes and from RNA sequences across multiple tissues. Approximately 60% of genes were the core genome (found in all species), and while synteny was mostly conserved across the species, large scale inversions and translocations were seen within subclades. While gene content was largely consistent between species, genome sizes ranged from 0.7-2.5 Gb mainly due to retrotransposon activity, but also from gene duplications. Over half a million duplications were found, both whole genome and single-gene, and paralogues disproportionally evolved to maintain normal gene dosage. This was greatly achieved through conservation of the cis-regulatory sequences compared to the coding portion of genes through evolution. Paralogues with similar expression patterns may be partially or fully redundant, compensatory, or pseudogenized, which may mean related genes in another species have different functions and effects on plant phenotypes. This complicates genetic engineering for crop improvement, as the effect on phenotype in one species may not reliably predict the outcome in others. However, with the continual establishment of lineage-specific pan-genomes, combined with advances in machine learning models, enhancement in the prediction accuracy is anticipated, paving the way for more precise and efficient crop improvement strategies. (Summary by Ciara O’Brien @ciara.obrien.bsky.social) Nature 10.1038/s41586-025-08619-6


 Soybean, originally domesticated in China, is widely recognized for its high protein and oil content, making it a crucial crop for human consumption, animal feed, and biofuel production. Despite its economic and agricultural significance, research tools and genomic resources for soybean have lagged behind those of other model species. The first genome assembly of a cultivated soybean variety was published 15 years ago, followed by the wild soybean genome nearly a decade later. While multi-omics data for soybean have grown rapidly in recent years, a centralized and user-friendly platform for accessing and analyzing these datasets remains underdeveloped. To bridge this gap, Fan and colleagues have established SoyOmics (https://ngdc.cncb.ac.cn/soyomics/transcriptome), a comprehensive database integrating bulk RNA-seq data from 314 samples spanning the soybean life cycle. Additionally, it includes single-nucleus RNA sequencing (snRNA-seq) and Stereo-seq datasets from five key organs: root, nodule, shoot apical meristem, leaf, and stem. This platform provides an invaluable resource for the soybean research community, facilitating gene association mapping, network analysis, and functional studies to advance soybean biology and breeding efforts. (Summary by Ching Chan
Soybean, originally domesticated in China, is widely recognized for its high protein and oil content, making it a crucial crop for human consumption, animal feed, and biofuel production. Despite its economic and agricultural significance, research tools and genomic resources for soybean have lagged behind those of other model species. The first genome assembly of a cultivated soybean variety was published 15 years ago, followed by the wild soybean genome nearly a decade later. While multi-omics data for soybean have grown rapidly in recent years, a centralized and user-friendly platform for accessing and analyzing these datasets remains underdeveloped. To bridge this gap, Fan and colleagues have established SoyOmics (https://ngdc.cncb.ac.cn/soyomics/transcriptome), a comprehensive database integrating bulk RNA-seq data from 314 samples spanning the soybean life cycle. Additionally, it includes single-nucleus RNA sequencing (snRNA-seq) and Stereo-seq datasets from five key organs: root, nodule, shoot apical meristem, leaf, and stem. This platform provides an invaluable resource for the soybean research community, facilitating gene association mapping, network analysis, and functional studies to advance soybean biology and breeding efforts. (Summary by Ching Chan 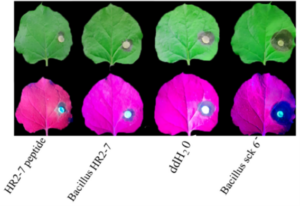 Microbial pesticides are widely applied to improve crop production, but the active molecules are largely unexplored. In a recent study, Mohamed et al., identified an antimicrobial peptide (AMP) from crop-associated microbes and showed its inhibitory effects on various phytopathogens. The authors first isolated bacterial strains from healthy crops and tested their antifungal activities. A Burkholderia strain highly inhibited the phytopathogen Rhizoctonia solani. A genomic library of the Burkholderia strain was constructed, and the antimicrobial effects of the genomic fragments were tested in Bacillus subtilis. Colony morphology and DNA sequence analyses revealed 29 AMPs, which were then tested for their antimicrobial effects. The 24aa AMP, namely HR2-7, weakened the pathogenicity of Botryosphaeria dothidea MAO, Dissotis theifolia CJP4-1, and Pseudomonas syringae DC 3000, which are pathogens of pear, tea, and tomato respectively. To explore the possible application of HR2-7 in the field, the authors tested its stability under different conditions including heat, UV exposure, and extreme pHs. Despite the sensitivity to UV exposure and extreme pHs, HR2-7 retained its antipathogenic effects at 60 °C. The thermostability favours its potential use in the field under global warming. (Summary by Yee-Shan Ku
Microbial pesticides are widely applied to improve crop production, but the active molecules are largely unexplored. In a recent study, Mohamed et al., identified an antimicrobial peptide (AMP) from crop-associated microbes and showed its inhibitory effects on various phytopathogens. The authors first isolated bacterial strains from healthy crops and tested their antifungal activities. A Burkholderia strain highly inhibited the phytopathogen Rhizoctonia solani. A genomic library of the Burkholderia strain was constructed, and the antimicrobial effects of the genomic fragments were tested in Bacillus subtilis. Colony morphology and DNA sequence analyses revealed 29 AMPs, which were then tested for their antimicrobial effects. The 24aa AMP, namely HR2-7, weakened the pathogenicity of Botryosphaeria dothidea MAO, Dissotis theifolia CJP4-1, and Pseudomonas syringae DC 3000, which are pathogens of pear, tea, and tomato respectively. To explore the possible application of HR2-7 in the field, the authors tested its stability under different conditions including heat, UV exposure, and extreme pHs. Despite the sensitivity to UV exposure and extreme pHs, HR2-7 retained its antipathogenic effects at 60 °C. The thermostability favours its potential use in the field under global warming. (Summary by Yee-Shan Ku 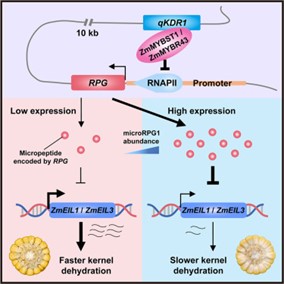 Kernel dehydration rate (KDR) is a critical factor affecting mechanized maize harvesting and kernel quality. Despite its agricultural importance, the molecular mechanisms regulating KDR remain unclear. Previous studies have identified several QTLs controlling KDR, but their functional characterization has been limited. Yu et al. investigate the genetic regulation of KDR by focusing on qKDR1, a major QTL that influences kernel moisture content at harvest. The study reveals that qKDR1 regulates the expression of qKDR1 REGULATED PEPTIDE GENE (RPG), which encodes microRPG1, a 31-amino acid Zea genus-specific micropeptide that originated de novo from a non-coding sequence. The authors demonstrate that microRPG1 modulates kernel dehydration by precisely regulating ZmETHYLENE-INSENSITIVE3-like 1 and 3 (ZmEIL1 and ZmEIL3), key components of the ethylene signaling pathway. Loss-of-function mutations in microRPG1 result in accelerated kernel dehydration, whereas overexpression or exogenous application of the micropeptide delays dehydration in both maize and Arabidopsis thaliana. This suggests that microRPG1 functions as a negative regulator of KDR by suppressing ethylene signaling. This study enhances our understanding of maize kernel dehydration and identifies microRPG1 as a key regulator of this process. Modulating its expression could help develop maize varieties with improved dehydration rates, benefiting mechanized harvesting and reducing post-harvest drying cost. (Summary by Muhammad Aamir Khan
Kernel dehydration rate (KDR) is a critical factor affecting mechanized maize harvesting and kernel quality. Despite its agricultural importance, the molecular mechanisms regulating KDR remain unclear. Previous studies have identified several QTLs controlling KDR, but their functional characterization has been limited. Yu et al. investigate the genetic regulation of KDR by focusing on qKDR1, a major QTL that influences kernel moisture content at harvest. The study reveals that qKDR1 regulates the expression of qKDR1 REGULATED PEPTIDE GENE (RPG), which encodes microRPG1, a 31-amino acid Zea genus-specific micropeptide that originated de novo from a non-coding sequence. The authors demonstrate that microRPG1 modulates kernel dehydration by precisely regulating ZmETHYLENE-INSENSITIVE3-like 1 and 3 (ZmEIL1 and ZmEIL3), key components of the ethylene signaling pathway. Loss-of-function mutations in microRPG1 result in accelerated kernel dehydration, whereas overexpression or exogenous application of the micropeptide delays dehydration in both maize and Arabidopsis thaliana. This suggests that microRPG1 functions as a negative regulator of KDR by suppressing ethylene signaling. This study enhances our understanding of maize kernel dehydration and identifies microRPG1 as a key regulator of this process. Modulating its expression could help develop maize varieties with improved dehydration rates, benefiting mechanized harvesting and reducing post-harvest drying cost. (Summary by Muhammad Aamir Khan 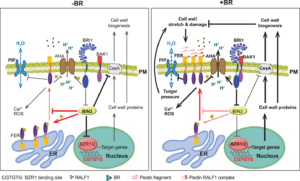 FERONIA (FER), a member of the Catharanthus roseus receptor-like kinase 1–like (CrRLK1-like) receptor kinase family, was initially identified as a key regulator of female fertility in Arabidopsis. Later studies revealed that FER is a multifunctional protein involved in nearly all aspects of plant growth and development, including cell proliferation, hormone signaling, reproduction, and responses to biotic and abiotic stresses. However, how a single receptor kinase mediates such diverse functions remains unclear. One approach to addressing this question is to investigate FER co-receptors and binding partners, which determine its downstream signaling pathways. Chaudhary and colleagues demonstrated that the interaction between BRASSINOSTEROID-INSENSITIVE 2 (BIN2) and FER plays a crucial role in brassinosteroid (BR)-induced cell expansion. In regions where cells are not actively proliferating or elongating, BR levels are low, allowing BIN2 to phosphorylate FER and suppress its localization on the plasma membrane. Conversely, when BR levels rise, BIN2 is inhibited, leading to FER accumulation on the plasma membrane. This, in turn, promotes reactive oxygen species (ROS) production, reduces cell wall acidification, slows down cell elongation, and helps maintain cell wall integrity during expansion. (Summary by Ching Chan
FERONIA (FER), a member of the Catharanthus roseus receptor-like kinase 1–like (CrRLK1-like) receptor kinase family, was initially identified as a key regulator of female fertility in Arabidopsis. Later studies revealed that FER is a multifunctional protein involved in nearly all aspects of plant growth and development, including cell proliferation, hormone signaling, reproduction, and responses to biotic and abiotic stresses. However, how a single receptor kinase mediates such diverse functions remains unclear. One approach to addressing this question is to investigate FER co-receptors and binding partners, which determine its downstream signaling pathways. Chaudhary and colleagues demonstrated that the interaction between BRASSINOSTEROID-INSENSITIVE 2 (BIN2) and FER plays a crucial role in brassinosteroid (BR)-induced cell expansion. In regions where cells are not actively proliferating or elongating, BR levels are low, allowing BIN2 to phosphorylate FER and suppress its localization on the plasma membrane. Conversely, when BR levels rise, BIN2 is inhibited, leading to FER accumulation on the plasma membrane. This, in turn, promotes reactive oxygen species (ROS) production, reduces cell wall acidification, slows down cell elongation, and helps maintain cell wall integrity during expansion. (Summary by Ching Chan 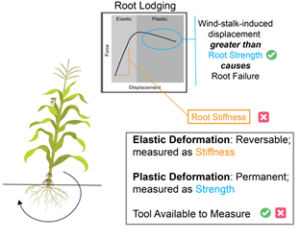 Field-based technologies are essential for understanding plant mechanical behavior in field-relevant contexts. This behavior can be elastic, with temporary deformation measured as stiffness, or plastic, with permanent deformation measured as strength. While field-based technologies exist for addressing questions regarding stalk stiffness, stalk strength, and root system strength, root system stiffness has not been fully understood – until now. A new study by Hostetler et al. introduces SMURF (Sorghum and Maize Under Rotational Force), a non-destructive field-based mechanical phenotyping device designed to measure root system resistance to bending and rotational force. This tool will enable researchers to assess root system stiffness in large grain crops. Putting the technology to test, the SMURF was used to measure root system stiffness in maize inbred and hybrid genotypes across developmental stages and varying planting densities. A key takeaway? A plant with a higher root system stiffness is more prone to mechanical failure under equal stalk displacement than a plant with lower root system stiffness. The SMURF opens new doors for understanding the complex relationship between root-soil interaction and plant mechanics. (Summary by Irene I. Ikiriko
Field-based technologies are essential for understanding plant mechanical behavior in field-relevant contexts. This behavior can be elastic, with temporary deformation measured as stiffness, or plastic, with permanent deformation measured as strength. While field-based technologies exist for addressing questions regarding stalk stiffness, stalk strength, and root system strength, root system stiffness has not been fully understood – until now. A new study by Hostetler et al. introduces SMURF (Sorghum and Maize Under Rotational Force), a non-destructive field-based mechanical phenotyping device designed to measure root system resistance to bending and rotational force. This tool will enable researchers to assess root system stiffness in large grain crops. Putting the technology to test, the SMURF was used to measure root system stiffness in maize inbred and hybrid genotypes across developmental stages and varying planting densities. A key takeaway? A plant with a higher root system stiffness is more prone to mechanical failure under equal stalk displacement than a plant with lower root system stiffness. The SMURF opens new doors for understanding the complex relationship between root-soil interaction and plant mechanics. (Summary by Irene I. Ikiriko 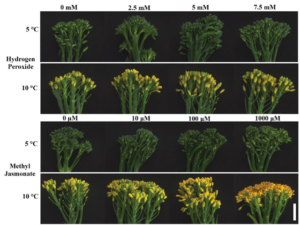 Up to 40% of harvested broccoli never reaches consumers due to quality losses during senescence, including yellowing and nutrient degradation. Reactive oxygen species cause oxidative stress within the plant, accelerating these negative changes through the jasmonate (JA) pathway. Gage et al. investigated if oxidative hormesis, where a mild activation of the stress-response pathway improves the plants’ ability to tolerate subsequent extreme stress, could help maintain the quality of Tenderstem® broccoli during postharvest storage. Despite success in other broccoli varieties, treatment with hydrogen peroxide did not maintain quality, and methyl JA exacerbated yellowing. This study also found that in Tenderstem®, early yellowing is due to carotenoid accumulation rather than chlorophyll degradation, differing from previous observations in other broccoli varieties. The authors suggest that the later developmental stage of Tenderstem® at harvest, along with genotypic differences, contributes to the altered response. In addition, this study also demonstrated that lower storage temperatures (5 vs. 10 °C) reduced JA-pathway gene expression, effectively prolonging green colour retention for over eight days. While chilling is energy-intensive, reducing food loss carries environmental and economic benefits, making it a worthwhile tradeoff. Overall, this study highlights the importance of understanding the diversity in stress responses and developmental differences between crop varieties, which impact postharvest strategies to minimize food losses. (Summary by Ciara O’Brien
Up to 40% of harvested broccoli never reaches consumers due to quality losses during senescence, including yellowing and nutrient degradation. Reactive oxygen species cause oxidative stress within the plant, accelerating these negative changes through the jasmonate (JA) pathway. Gage et al. investigated if oxidative hormesis, where a mild activation of the stress-response pathway improves the plants’ ability to tolerate subsequent extreme stress, could help maintain the quality of Tenderstem® broccoli during postharvest storage. Despite success in other broccoli varieties, treatment with hydrogen peroxide did not maintain quality, and methyl JA exacerbated yellowing. This study also found that in Tenderstem®, early yellowing is due to carotenoid accumulation rather than chlorophyll degradation, differing from previous observations in other broccoli varieties. The authors suggest that the later developmental stage of Tenderstem® at harvest, along with genotypic differences, contributes to the altered response. In addition, this study also demonstrated that lower storage temperatures (5 vs. 10 °C) reduced JA-pathway gene expression, effectively prolonging green colour retention for over eight days. While chilling is energy-intensive, reducing food loss carries environmental and economic benefits, making it a worthwhile tradeoff. Overall, this study highlights the importance of understanding the diversity in stress responses and developmental differences between crop varieties, which impact postharvest strategies to minimize food losses. (Summary by Ciara O’Brien