Mathematical modeling approaches in plant metabolomics
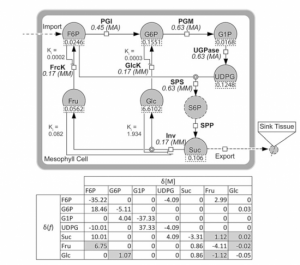 Mathematical modeling, as a series of useful methods for functional and causal interpretation of experimental high-throughput studies, enables us to explore the complex field of plant metabolism. The application of differential equations as a mathematical method has been developed and reviewed for decades and has been used to describe, analyze, and predict complex and nonintuitive system dynamics in plant biochemistry and physiology. Fittingly to the limited availability of kinetic information required, alternative approaches have been developed, e.g. system linearization, and parameter normalization. The quasi steady state Jacobian matrices are consequently introduced as an instrument for connecting metabolite concentrations and/or enzyme activities with biochemical network information. Even though statistical methods are an invaluable tool, it is recommended to combine them with biochemical information in order to enhance the reliability and predictive power of the output. In the context of metabolomic studies, this biochemical information can typically be derived from a genome-scale metabolic network. (Summary by Tina Ugulin) Fürtauer L., Weiszmann J., Weckwerth W., Nägele T. (2018) Mathematical Modeling Approaches in Plant Metabolomics. In: António C. (eds) Plant Metabolomics. Methods in Molecular Biology, vol 1778. Humana Press, New York, NY
Mathematical modeling, as a series of useful methods for functional and causal interpretation of experimental high-throughput studies, enables us to explore the complex field of plant metabolism. The application of differential equations as a mathematical method has been developed and reviewed for decades and has been used to describe, analyze, and predict complex and nonintuitive system dynamics in plant biochemistry and physiology. Fittingly to the limited availability of kinetic information required, alternative approaches have been developed, e.g. system linearization, and parameter normalization. The quasi steady state Jacobian matrices are consequently introduced as an instrument for connecting metabolite concentrations and/or enzyme activities with biochemical network information. Even though statistical methods are an invaluable tool, it is recommended to combine them with biochemical information in order to enhance the reliability and predictive power of the output. In the context of metabolomic studies, this biochemical information can typically be derived from a genome-scale metabolic network. (Summary by Tina Ugulin) Fürtauer L., Weiszmann J., Weckwerth W., Nägele T. (2018) Mathematical Modeling Approaches in Plant Metabolomics. In: António C. (eds) Plant Metabolomics. Methods in Molecular Biology, vol 1778. Humana Press, New York, NY



