Is Genetic Evolution Predictable?
How often does evolution repeat itself? When the same evolutionary strategy arises multiple times, how often are these strategies built on the same genetic foundations? Addressing this question allows us to understand the relative roles of constraint and contingency in the history of life, but (without a time machine) identifying independent and parallel evolutionary strategies requires careful observation and experimentation.
Flowering during appropriate conditions is key to adaptation, if you are an angiosperm. Capsella rubella experienced a bottleneck during its recent origin (Foxe et al. 2009; Guo et al. 2009), so a lack of standing genetic diversity could plausibly limit the evolution of novel flowering time phenotypes. Despite this, natural accessions of C. rubella display wide variation for flowering time (Guo et al., 2012). To understand how, Yang et al. (2018) undertook a series of thoughtful experiments which narrow in on a twice-evolved genetic mechanism underlying earlier flowering time.
Yang and colleagues began by generating two F2 mapping populations, crossing the late-flowering reference accession MTE with two earlier flowering accessions, 86IT1 and 762. They identified a genomic region in both populations enriched for allele frequency differences in early flowering F2 plants that contains the C. rubella ortholog of FLC (CrFLC), a cental regulator of flowering time (Sheldon et al. 2000). Sequencing the CrFLC locus in the three parental accessions, Yang and colleagues found overlapping 5’ UTR deletions in both earlier flowering parents relative to the later flowering MTE (see Figure). The two parents with a 5’ UTR deletion CrFLC allele (i.e. deletion allele) express lower levels of CrFLC and are less responsive to vernalization, during which FLC expression is typically repressed (Sheldon et al. 2000).
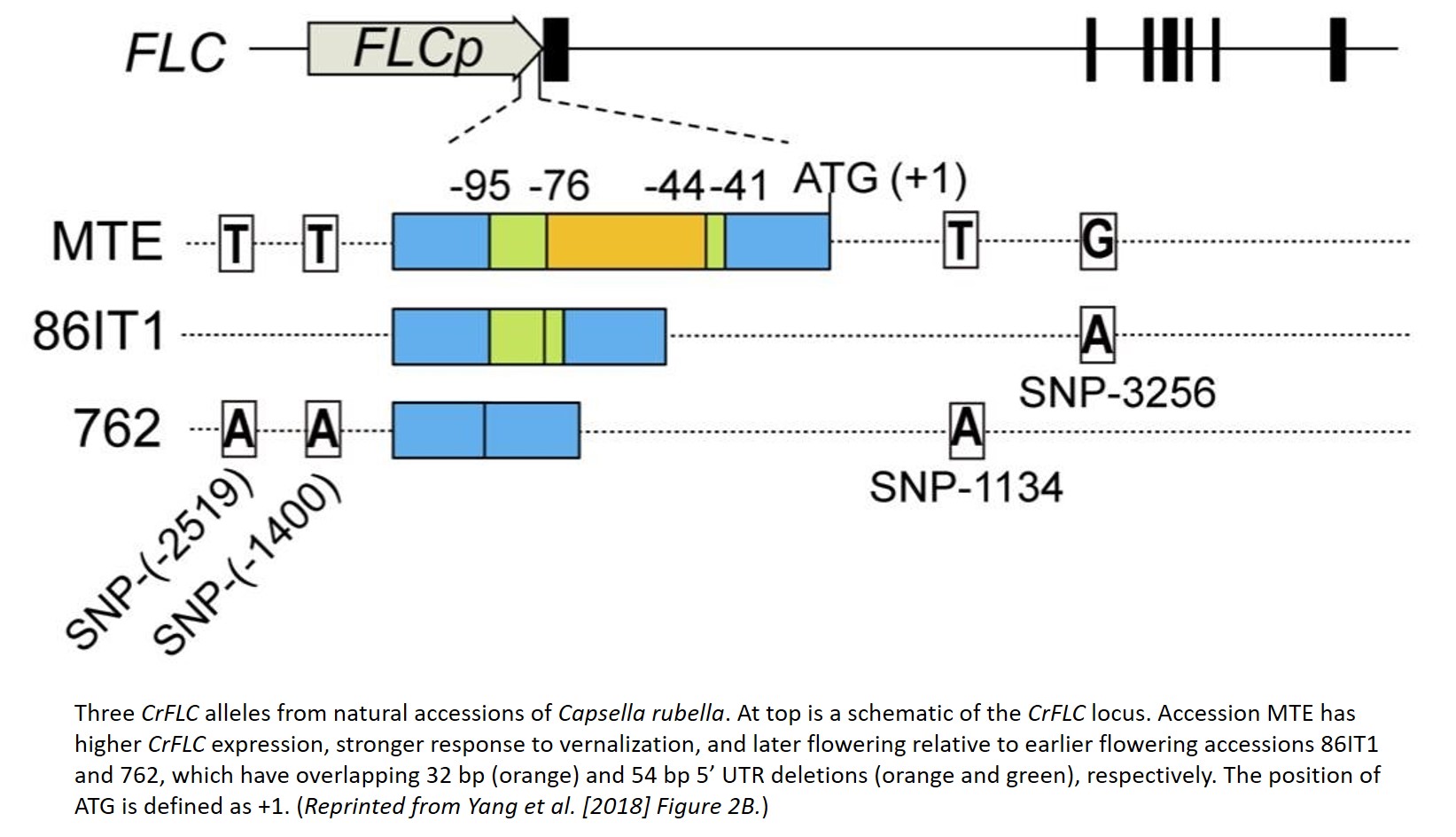
When transformed into A. thaliana, the CrFLC deletion alleles continue to confer lower levels of FLC expression and earlier flowering than the MTE allele. These transformed alleles also affect histone modification: the MTE allele is enriched for signals of active transcription, and the deletion alleles are enriched for repressive chromatin marks, with these differences most pronounced in the 5’ UTR. Deleting a section from the 5’ UTR of the MTE allele creates an expression and flowering phenotype similar to the natural deletion alleles, and re-introducing the deleted region to a deletion allele creates a phenotype similar to the MTE allele. Through these experiments and several others, Yang and colleagues show that a 5’ UTR deletion is sufficient to reduce the expression of CrFLC and induce earlier flowering, likely through changes in histone modification and chromatin structure.
With mechanism in hand, Yang and colleagues confirm that the CrFLC deletion alleles have arisen independently and are segregating in natural accessions of C. rubella. In these accessions, CrFLC 5’ UTR genotype is associated with both changes in CrFLC expression and flowering time. Although we cannot (yet) “replay the tape” of life, studies like this one show that sometimes life finds a way… to repeat itself.
REFERENCES
Foxe, J.P., Slotte, T., Stahl, E.A., Neuffer, B., Hurka, H., & Wright, S.I. (2009). Recent speciation associated with the evolution of selfing in Capsella. PNAS 106: 5241-5245.
Guo, Y.L., Todesco, M., Hagmann, J., Das, S., & Weigel, D. (2012). Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics 192: 729-739.
Guo, Y.L., Bechsgaard, J.S., Slotte, T., Neuffer, B., Lascoux, M., Weigel, D., & Schierup, M.H. (2009). Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. PNAS 106: 5246-5251.
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., & Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). PNAS 97: 3753-3758.
Yang, L., Wang, H.N., Hou, X.H., Zou, Y.P., Han, T.S., Niu, X.M., Zhang, J., Zhao, Z., Todesco, M., Balasubramanian, S., & Guo, Y.L. (2018). Parallel evolution of common allelic variants confers flowering diversity in Capsella rubella. Plant Cell. DOI:10.1105/tpc.18.00124.