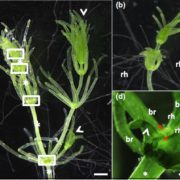
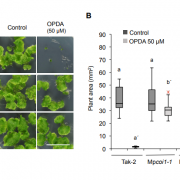
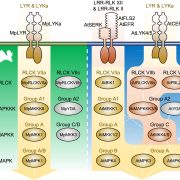
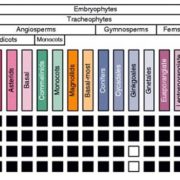
Genome editing retraces the evolution of toxin resistance in the monarch butterfly ($) (Nature)
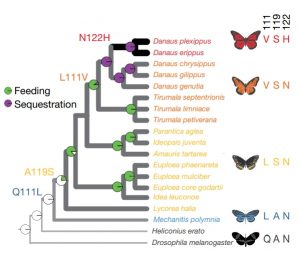 When a few species from several distantly related groups produce a similar but unusual trait, we usually assume that this trait is an example of convergent evolution; starting from different places but ending up at the same place. The ability to eat plants that produce cardiac glycosides, which are toxic to most animals, occurs in a few species from across six insect orders, so is considered an example of convergent evolution. Thanks to genome sequencing and the ability to edit genomes through CRISPR/Cas9, it is now possible to directly test these assumptions. Karageorgi et al. have done that, by identifying key substitutions in ATPα that occur in multiple resistant species, and also determining the apparent order in which they arose. The authors then introduced these substitutions into Drosophila and found as the increased the number of candidate substitutions, resistance to ouabain (a cardiac glycoside) increased. The authors conclude that this is “the first in vivo validation of a multi-step adaptive walk in a multicellular organism.” (Summary by Mary Williams) Nature 10.1038/s41586-019-1610-8
When a few species from several distantly related groups produce a similar but unusual trait, we usually assume that this trait is an example of convergent evolution; starting from different places but ending up at the same place. The ability to eat plants that produce cardiac glycosides, which are toxic to most animals, occurs in a few species from across six insect orders, so is considered an example of convergent evolution. Thanks to genome sequencing and the ability to edit genomes through CRISPR/Cas9, it is now possible to directly test these assumptions. Karageorgi et al. have done that, by identifying key substitutions in ATPα that occur in multiple resistant species, and also determining the apparent order in which they arose. The authors then introduced these substitutions into Drosophila and found as the increased the number of candidate substitutions, resistance to ouabain (a cardiac glycoside) increased. The authors conclude that this is “the first in vivo validation of a multi-step adaptive walk in a multicellular organism.” (Summary by Mary Williams) Nature 10.1038/s41586-019-1610-8