Fishing out virus replication complexes
Incarbone et al. use an epitope-tagged protein that binds double-stranded RNAs to fish out viral replication complexes and identify the associated proteins. Plant Cell. https://doi.org/10.1093/plcell/koab214
Marco Incarbone (GMI, Vienna) and Christophe Ritzenthaler (IBMP, Strasbourg)
Background: Viruses can cause devastating diseases in plants, and understanding their molecular workings is of great importance. All RNA viruses, which constitute the majority of plant virus species, generate double-stranded RNA (dsRNA) during their replication process. Replication occurs within so-called virus replication complexes (VRCs), which are highly dynamic macro-molecular machines composed of both virus- and host-encoded proteins and are housed within ad hoc intracellular niches. The experimental tools available to investigate the molecular composition of VRCs are currently limited to specific recombinant virus species or to surrogate hosts such as yeast. However, we lacked a technique to reliably and easily isolate replicating viral double-stranded RNA and identify the associated proteins.
Question: We wondered whether we could use an epitope-tagged double-stranded RNA-binding protein to isolate replicating viral RNA. If this could be achieved, we wondered whether we could identify by mass spectrometry any proteins associated with the double-stranded RNA and confirm their localization at VRCs through fluorescence microscopy.
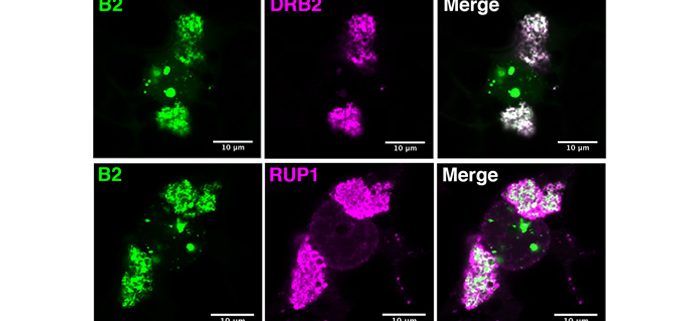
Findings: After generating Arabidopsis thaliana lines expressing the flock house virus-encoded dsRNA-binding domain of protein B2 fused to GFP, we confirmed its localization at VRCs during tobacco rattle virus infection. Through GFP immunoprecipitation we were able to isolate viral double-stranded RNA and, through mass spectrometry, identify a number of potentially associated proteins, including the viral replicase enzyme. We could confirm by microscopy that several of these host-encoded proteins localize at or near VRCs during infection. Furthermore, one of these proteins – DRB2 – showed remarkable antiviral activity against several RNA virus species.
Next steps: Next steps should involve the development and adaptation of this B2-based system to isolate replicating RNA of other viruses in other plant species and possibly also under native conditions. Importantly, this system should be further adapted to investigate the replication complexes of human and animal RNA viruses. Additional work on DRB2 as an antiviral biotech tool also holds great potential.
Reference:
Marco Incarbone, Marion Clavel, Baptiste Monsion, Lauriane Kuhn, Hélène Scheer, Émilie Vantard, Vianney Poignavent, Patrice Dunoyer, Pascal Genschik, Christophe Ritzenthaler (2021) Immunocapture of dsRNA-bound proteins provides insight into Tobacco rattle virus replication complexes and reveals Arabidopsis DRB2 to be a wide-spectrum antiviral effector. https://doi.org/10.1093/plcell/koab214