Antagonistic regulation of tomato branching
Wang et al. revealed fine regulation of inflorescence branching in tomato. The Plant Cell (2023).
By X.T. Wang and X. Cui
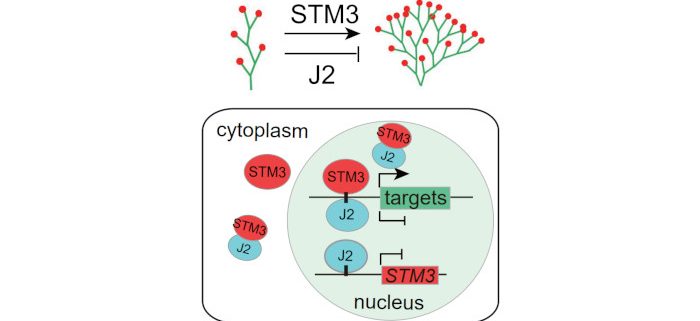
Background: The number of branches in the inflorescence affects yield in food crops and the aesthetics of ornamental plants. Two MADS-box transcription factors, SISTER OF TM3 (STM3) and JOINTLESS2 (J2), are tomato homologs of Arabidopsis SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 and SEPALLATA 4, respectively, and have opposing regulatory functions in controlling inflorescence branching.
Question: How do STM3 and J2 antagonistically regulate inflorescence branching in tomato?
Findings: We used chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) and electrophoretic mobility shift assays (EMSA) to identify and confirm a set of common putative target genes that are directly bound by STM3 and J2 via the CArG box motif in their promoter regions. One of these direct targets, FRUITFULL1 (FUL1), is antagonistically regulated by STM3 and J2 during inflorescence development. STM3 forms a complex with J2 and mediates its translocation from the cytosol to the nucleus, thus restricting J2 repressor activity. J2 also limits STM3 function by repressing STM3 expression and inhibits its binding activity. Our results suggest an antagonistic regulatory mode in which STM3 and J2 exert flexible control over inflorescence meristem determinacy and branch number.
Next steps: We and others aim to identify additional factors affecting tomato inflorescence development and the environmental adaptability of inflorescence branching.
Reference:
Xiaotian Wang, Zhiqiang Liu, Jingwei Bai, Shuai Sun, Jia Song, Ren Li, Xia Cui. (2023). Antagonistic regulation of target genes by the SISTER OF TM3–JOINTLESS2 complex in tomato inflorescence branching. https://doi.org/10.1093/plcell/koad065