97 Shades of Gray: Genetic interactions of the gray mold, Botrytis cinerea, with wild and domesticated tomato
Although a vineyard full of decaying grapes infected with noble rot is a blessing for sweet wine producers, the causal agent, Botrytis cinerea (gray mold), causes huge crop losses. Unlike most plant pathogens, individual isolates of the necrotrophic fungus can infect an extremely broad range of plants, including cereals, fruits, vegetables, and ornamentals. Host immune responses to such generalist pathogens are complex, involving multiple defense mechanisms and pathways with resistance to B. cinerea segregating genetically as a quantitative trait (Rowe and Kliebenstein, 2008). Little is known, however, about the intricacies of this host-pathogen genetic interaction.
A new study by Soltis et al. (2019) utilizes the tomato-B. cinerea pathosystem to measure how genetic diversity in both the host plant and a generalist pathogen interact to affect resistance/susceptibility, but on a previously unimaginable scale. They measured 1164 unique plant x pathogen genetic interactions by inoculating 97 genetically distinct B. cinerea isolates on six domesticated (Solanum lycopersicum) and six wild (S. pimpinellifolium) tomato accessions, totaling 3276 independent necrotic lesion areas.
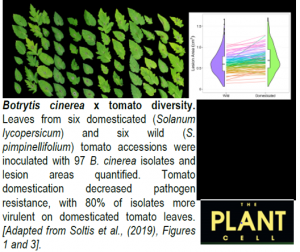 Specialist pathogens that infect a very narrow host range frequently have only a handful of large-effect qualitative loci controlling virulence. By contrast, genome-wide association (GWA) mapping across the 97 isolates revealed that B. cinerea virulence on tomato is extremely polygenic, with numerous small-effect single nucleotide polymorphisms (SNPs) linked to differential virulence. This is directly akin to the polygenic basis of resistance in the host plant (Zhang et al., 2017). Furthermore, most of the identified SNPs were only linked to a subset of tomato accessions, demonstrating that B. cinerea virulence is highly dependent on host genotype. These SNPs were associated with several hundred B. cinerea genes, including both previously characterized and novel virulence loci. Detailed examination of multiple SNPs within one of these, a pectinesterase gene, revealed that most were located in upstream regulatory regions. Subtleties in the transcriptional regulation of virulence-related proteins may therefore exist between individual B. cinerea isolates.
Specialist pathogens that infect a very narrow host range frequently have only a handful of large-effect qualitative loci controlling virulence. By contrast, genome-wide association (GWA) mapping across the 97 isolates revealed that B. cinerea virulence on tomato is extremely polygenic, with numerous small-effect single nucleotide polymorphisms (SNPs) linked to differential virulence. This is directly akin to the polygenic basis of resistance in the host plant (Zhang et al., 2017). Furthermore, most of the identified SNPs were only linked to a subset of tomato accessions, demonstrating that B. cinerea virulence is highly dependent on host genotype. These SNPs were associated with several hundred B. cinerea genes, including both previously characterized and novel virulence loci. Detailed examination of multiple SNPs within one of these, a pectinesterase gene, revealed that most were located in upstream regulatory regions. Subtleties in the transcriptional regulation of virulence-related proteins may therefore exist between individual B. cinerea isolates.
Taken together, these findings truly highlight the tenacity of B. cinerea as a plant pathogen! The high level of standing genetic diversity found in B. cinerea virulence loci, coupled with the ability of different isolates to intermate, means that the pathogen has at its disposal a cornucopia of beneficial alleles. This not only allows selection to customize virulence according to host genotype, but also enables rapid adaptation to any newly evolved plant defense mechanisms or host niches. Thus, breeding durable resistance to B. cinerea in the field is extremely challenging and will likely require a genome-wide appreciation of virulence/resistance at both the pathogen and host levels.
REFERENCES
Rowe, H.C. and Kliebenstein, D.J. (2008). Complex Genetics Control Natural Variation in Arabidopsis thaliana Resistance to Botrytis cinerea. Genetics 180: 2237–2250.
Soltis, N.E., Atwell, S., Shi, G., Fordyce, R., Gwinner, R., Gao, D., Shafi, A., and Kliebenstein, D.J. (2019). Interactions of tomato and Botrytis genetic diversity: Parsing the contributions of host differentiation, domestication and pathogen variation. Plant Cell 31: doi:10.1015/tpc18.00857.
Zhang, W., Corwin, J.A., Copeland, D., Feusier, J., Eshbaugh, R., Chen, F., Atwell, S., and Kliebenstein, D.J. (2017). Plastic Transcriptomes Stabilize Immunity to Pathogen Diversity: The Jasmonic Acid and Salicylic Acid Networks within the Arabidopsis/ Botrytis Pathosystem. Plant Cell 29: 2727–2752.



