What We’re Reading: May 26th
Review: Enhancing genetic gain in the era of molecular breeding ($)
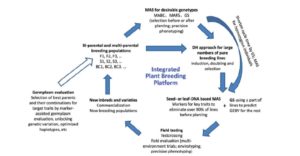 Yield is determined by the crop’s genetic potential and the realization of that potential as affected by agronomic practices and environmental factors. Xu et al. address how yields can be improved through enhancing genetic gain, which they define as “the amount of increase in performance that is achieved annually through artificial selection.” Factors that affect genetic gain include the genetic variance of the population (including variance that can be created as well as contributions from wild relatives), heritability, selection intensity (affected by methods used to assess phenotype and their costs), and cycle time (which can be accelerated through greenhouses and winter nurseries). This review, recommended reading for students, nicely demonstrates how, “Advances in biological science and technology, which involve genomics, informatics, modeling, communication technologies, satellite imaging, remote sensing, and precision farming and agriculture, have contributed significantly to increasing crop productivity.” J. Exp. Bot. 10.1093/jxb/erx135 Tags: Applied Plant Biology, Food Security, Genetics
Yield is determined by the crop’s genetic potential and the realization of that potential as affected by agronomic practices and environmental factors. Xu et al. address how yields can be improved through enhancing genetic gain, which they define as “the amount of increase in performance that is achieved annually through artificial selection.” Factors that affect genetic gain include the genetic variance of the population (including variance that can be created as well as contributions from wild relatives), heritability, selection intensity (affected by methods used to assess phenotype and their costs), and cycle time (which can be accelerated through greenhouses and winter nurseries). This review, recommended reading for students, nicely demonstrates how, “Advances in biological science and technology, which involve genomics, informatics, modeling, communication technologies, satellite imaging, remote sensing, and precision farming and agriculture, have contributed significantly to increasing crop productivity.” J. Exp. Bot. 10.1093/jxb/erx135 Tags: Applied Plant Biology, Food Security, Genetics
Review: Systems-wide understanding of photosynthetic acclimation in algae and higher plants ($)
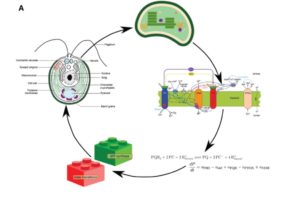 Deriving energy from light is a bit like keeping your hands warm by juggling burning coals; doable but dangerous. Photosynthetic organisms constantly adjust their photosynthetic machinery to optimize energy production but avoid damage from excess light. Moejes et al. describe a large-scale project, AccliPhot, that combined theoretical and experimental approaches to understand photosynthetic acclimation in algae and higher plants. They also describe progress towards using these insights to improve algal culture systems to optimize lipid production for biofuels. J. Exp. Bot. 10.1093/jxb/erx137 Tags: Applied Plant Biology, Bioenergy, Metabolism, Systems Biology
Deriving energy from light is a bit like keeping your hands warm by juggling burning coals; doable but dangerous. Photosynthetic organisms constantly adjust their photosynthetic machinery to optimize energy production but avoid damage from excess light. Moejes et al. describe a large-scale project, AccliPhot, that combined theoretical and experimental approaches to understand photosynthetic acclimation in algae and higher plants. They also describe progress towards using these insights to improve algal culture systems to optimize lipid production for biofuels. J. Exp. Bot. 10.1093/jxb/erx137 Tags: Applied Plant Biology, Bioenergy, Metabolism, Systems Biology
Insights from 292 pigeonpea genomes ($)
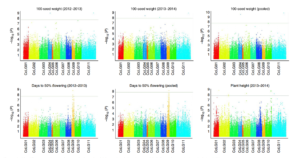 Pigeonpea (Cajanus cajan) is a widely-grown tropical legume domesticated in India about 3500 years ago. Yields have plateaued, and there is evidence for a genetic bottleneck that limits breeding potential in the elite varieties. Varshney et al. resequenced nearly 300 pigeonpea accessions including modern cultivars, traditional landraces and wild species. From these data they identified more than 17 million variants, including many regions that show evidence of selection during domestication and breeding. Marker-trait associations for flowering time and seed yield also were identified. Nature Genet. 10.1038/ng.3872 Tags: Genomics, Food Security
Pigeonpea (Cajanus cajan) is a widely-grown tropical legume domesticated in India about 3500 years ago. Yields have plateaued, and there is evidence for a genetic bottleneck that limits breeding potential in the elite varieties. Varshney et al. resequenced nearly 300 pigeonpea accessions including modern cultivars, traditional landraces and wild species. From these data they identified more than 17 million variants, including many regions that show evidence of selection during domestication and breeding. Marker-trait associations for flowering time and seed yield also were identified. Nature Genet. 10.1038/ng.3872 Tags: Genomics, Food Security
The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution
 Sunflower (Helianthus annuus L.) is an important oil crop and, according to the authors, “the only major crop domesticated in North America.” Assembling its genome has been difficult as more than three quarters of it is made up of young (less than one million years old) long-terminal repeat retrotransposons. Badouin et al. report a reference genome for sunflower which they generated from long reads. Their data revealed the evolutionary history of sunflower. Through quantitative genetics, and expression and diversity data, they identified genes and gene networks associated with flowering time and oil production, two important breeding traits. Nature 10.1038/nature22380 Tags: Genomics
Sunflower (Helianthus annuus L.) is an important oil crop and, according to the authors, “the only major crop domesticated in North America.” Assembling its genome has been difficult as more than three quarters of it is made up of young (less than one million years old) long-terminal repeat retrotransposons. Badouin et al. report a reference genome for sunflower which they generated from long reads. Their data revealed the evolutionary history of sunflower. Through quantitative genetics, and expression and diversity data, they identified genes and gene networks associated with flowering time and oil production, two important breeding traits. Nature 10.1038/nature22380 Tags: Genomics
Arabidopsis proteins with a transposon-related domain act in gene silencing
 While screening for mutants in gene silencing, Ikeda et al. identified mail1, which shows elevated expression of transposable elements (TEs) and protein-coding genes, indicating that MAIL1 (MAINTENANCE OF MERISTEM-LIKE1) is required for epigenetic silencing of some genes. However, mail1 does not differ from wildtype in either DNA methylation or siRNA accumulation, indicating it is involved in a distinct form of gene silencing. DAPI staining showed that pericentromeric regions are decondensed in mail1. MAIL1 has an aminotransferase-like, plant mobile domain that is similar to a DNA-binding domain found in transposases. The function of this domain and whether it originated in a gene or a transposon remain unknown. Nature Comms. 10.1038/ncomms15122 Tags: Gene Regulation, Molecular Biology
While screening for mutants in gene silencing, Ikeda et al. identified mail1, which shows elevated expression of transposable elements (TEs) and protein-coding genes, indicating that MAIL1 (MAINTENANCE OF MERISTEM-LIKE1) is required for epigenetic silencing of some genes. However, mail1 does not differ from wildtype in either DNA methylation or siRNA accumulation, indicating it is involved in a distinct form of gene silencing. DAPI staining showed that pericentromeric regions are decondensed in mail1. MAIL1 has an aminotransferase-like, plant mobile domain that is similar to a DNA-binding domain found in transposases. The function of this domain and whether it originated in a gene or a transposon remain unknown. Nature Comms. 10.1038/ncomms15122 Tags: Gene Regulation, Molecular Biology
A multi-purpose toolkit to enable advanced genome engineering in plants
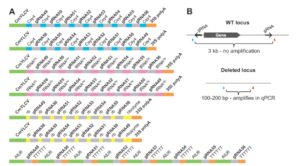 Precise genome editing holds tremendous promise for meeting future food security needs and sustainable agriculture. Čermák et al. describe a set of reagents that facilitate genome editing in plants, based on both TALEN (Transcription Activator-Like Effector Nucleases) and CRISPR/Cas technologies. Precise targeting of enzymes to specific regions of genomes enables many different types of editing. In this paper, the authors describe a “comprehensive, modular system for plant genome engineering that makes it possible to accomplish gene knockouts, replacements, altered transcriptional regulation or multiplexed modification.” Furthermore, these reagents have proven useful in both monocots and dicots. Detailed protocols are available in the Supplemental Information, and a website is available to select vectors and design primers. [The development and application of these reagents was recently mentioned in Science in a profile of the senior author, Daniel Voytas ($)]. Plant Cell 10.1105/tpc.16.00922 Tags: Genetics, Genomics, Molecular Biology
Precise genome editing holds tremendous promise for meeting future food security needs and sustainable agriculture. Čermák et al. describe a set of reagents that facilitate genome editing in plants, based on both TALEN (Transcription Activator-Like Effector Nucleases) and CRISPR/Cas technologies. Precise targeting of enzymes to specific regions of genomes enables many different types of editing. In this paper, the authors describe a “comprehensive, modular system for plant genome engineering that makes it possible to accomplish gene knockouts, replacements, altered transcriptional regulation or multiplexed modification.” Furthermore, these reagents have proven useful in both monocots and dicots. Detailed protocols are available in the Supplemental Information, and a website is available to select vectors and design primers. [The development and application of these reagents was recently mentioned in Science in a profile of the senior author, Daniel Voytas ($)]. Plant Cell 10.1105/tpc.16.00922 Tags: Genetics, Genomics, Molecular Biology
Target RNA secondary structure is a major determinant of miR159 efficacy
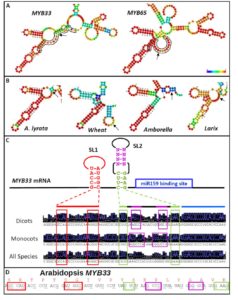 The interactions with microRNAs (miRNAs) and their targets in plants is assumed to be dependent largely on complementarity between the two RNAs. Zheng et al. investigated the interactions between miR159 and its putative targets, mRNA of several MYB genes, each with similar complementarity to the miRNA. Genetic data however suggest that miR159 silences some of these MYB genes more effectively than others. The authors found that the presence of two stem-loop RNA structures adjacent to the miRNA target site enhanced the efficacy of miR159. The authors suggest that the function of the stem-loop structures might be to increase the likelihood of the target site being single-stranded, or might facilitate interactions with proteins that would affect silencing efficacy. These results show that sequence complementarity alone is not sufficient to determine the efficacy of miRNAs, but that flanking sequences and structures are also important. Plant Physiol. Tags: Gene Regulation, Molecular Biology
The interactions with microRNAs (miRNAs) and their targets in plants is assumed to be dependent largely on complementarity between the two RNAs. Zheng et al. investigated the interactions between miR159 and its putative targets, mRNA of several MYB genes, each with similar complementarity to the miRNA. Genetic data however suggest that miR159 silences some of these MYB genes more effectively than others. The authors found that the presence of two stem-loop RNA structures adjacent to the miRNA target site enhanced the efficacy of miR159. The authors suggest that the function of the stem-loop structures might be to increase the likelihood of the target site being single-stranded, or might facilitate interactions with proteins that would affect silencing efficacy. These results show that sequence complementarity alone is not sufficient to determine the efficacy of miRNAs, but that flanking sequences and structures are also important. Plant Physiol. Tags: Gene Regulation, Molecular Biology
Towards an understanding of spiral patterning in the Sargassum muticum shoot apex
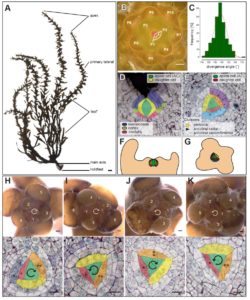 Classic studies of leaf placement (phyllotaxy) showed that two different mechanisms occur in land plants. In mosses, leaf placement is determined by the plane of cell division within the single apical cell, whereas in seed plants auxin acts as a morphogen that specifies leaf positioning. Linardić and Braybrook examined how the spiral pattern of phyllotaxy is determined in the brown alga Sargassum muticum. Although multicellularity arose independently in brown algae and plants, there are similarities in their indeterminate structure and apical meristem-like regions. The authors showed that in the brown alga the spiral orientation of the apical cell cleavage is unlinked to the spiral orientation of phyllotaxy. Destruction of the apical cell was frequently followed by reorganization and re-establishment of phyllotaxy, suggesting that it is a self-organizing principle, but there is no evidence for auxin involvement. BioRxiv 10.1101/135806 Tags: Botany, Development
Classic studies of leaf placement (phyllotaxy) showed that two different mechanisms occur in land plants. In mosses, leaf placement is determined by the plane of cell division within the single apical cell, whereas in seed plants auxin acts as a morphogen that specifies leaf positioning. Linardić and Braybrook examined how the spiral pattern of phyllotaxy is determined in the brown alga Sargassum muticum. Although multicellularity arose independently in brown algae and plants, there are similarities in their indeterminate structure and apical meristem-like regions. The authors showed that in the brown alga the spiral orientation of the apical cell cleavage is unlinked to the spiral orientation of phyllotaxy. Destruction of the apical cell was frequently followed by reorganization and re-establishment of phyllotaxy, suggesting that it is a self-organizing principle, but there is no evidence for auxin involvement. BioRxiv 10.1101/135806 Tags: Botany, Development
Bacterial biosensors for in vivo spatiotemporal mapping of root secretion
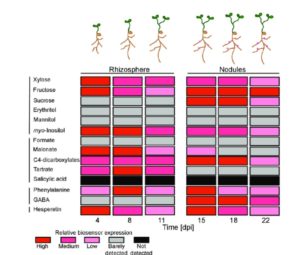 Biosensors are powerful tools that provide readouts for various small molecules so that they can be detected and located. Pini et al. have developed a set of biosensors for expression in bacteria (Rhizobium leguminosarum) that reveal some of the small molecules (including key sugars, polyols, organic acids, amino acids and flavonoids) secreted by the roots of its symbiotic partner pea (Pisum sativum). The authors developed biosensors by fusing the promoter element of a gene specifically expressed by a small molecule to the luciferase reporter lux. Nine of the 14 biosensors only responded to a single compound. The biosensors allowed the authors to spatially and temporally track rhizosphere metabolites during nodulation and to evaluate how different mutant backgrounds affected these metabolites. Plant Physiol. Tags: Biotic Interactions, Metabolism
Biosensors are powerful tools that provide readouts for various small molecules so that they can be detected and located. Pini et al. have developed a set of biosensors for expression in bacteria (Rhizobium leguminosarum) that reveal some of the small molecules (including key sugars, polyols, organic acids, amino acids and flavonoids) secreted by the roots of its symbiotic partner pea (Pisum sativum). The authors developed biosensors by fusing the promoter element of a gene specifically expressed by a small molecule to the luciferase reporter lux. Nine of the 14 biosensors only responded to a single compound. The biosensors allowed the authors to spatially and temporally track rhizosphere metabolites during nodulation and to evaluate how different mutant backgrounds affected these metabolites. Plant Physiol. Tags: Biotic Interactions, Metabolism
The RxLR motif of the Phytophthora infestans effector AVR3a is cleaved before secretion ($)
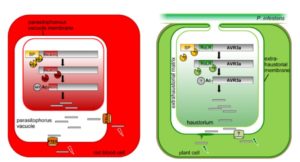 RxLR effectors are proteins secreted from pathogens that enter the cells of the host and support the effectiveness of the pathogen in various ways. Their name refers to the sequence RxLR (Arg-Xxx-Leu-Arg). Previously, this sequence has been thought to be involved the effector’s entry into the host cell. Wawra et al. used mass spectrometry and other approaches to examine the AVR3a effector secreted from cultured Phytophthora infestans. Their data show that the effector is cleaved at the RxLR site, and the resulting protein probably acetylated at the new N-terminus. This processing is similar to the that which occurs at the PEXEL (plasmodium export element) site of effectors of the Plasmodium pathogen that causes malaria. These new data argue that the RxLR sequence is processed within the pathogen cell and therefore not directly involved in uptake by the host cell. Plant Cell 10.1105/tpc.16.00552 Tags: Biotic Interactions, Cell Biology
RxLR effectors are proteins secreted from pathogens that enter the cells of the host and support the effectiveness of the pathogen in various ways. Their name refers to the sequence RxLR (Arg-Xxx-Leu-Arg). Previously, this sequence has been thought to be involved the effector’s entry into the host cell. Wawra et al. used mass spectrometry and other approaches to examine the AVR3a effector secreted from cultured Phytophthora infestans. Their data show that the effector is cleaved at the RxLR site, and the resulting protein probably acetylated at the new N-terminus. This processing is similar to the that which occurs at the PEXEL (plasmodium export element) site of effectors of the Plasmodium pathogen that causes malaria. These new data argue that the RxLR sequence is processed within the pathogen cell and therefore not directly involved in uptake by the host cell. Plant Cell 10.1105/tpc.16.00552 Tags: Biotic Interactions, Cell Biology
Global translational reprogramming of plant immune response, and engineering of disease resistance through regulated translation ($)
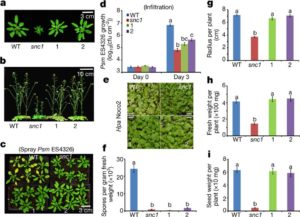 Plants fight back when pathogens attack, and in the first of a pair of papers Xu et al. have revealed a new insight into this response by demonstrating a significant translational upregulation of many defense-response mRNAs (10.1038/nature22371). Previously, this group showed that the translation of a key regulator of transcriptional responses to pathogens, TBF1, is itself regulated by the presence of two upstream open reading frames (uORFs). In the first of the new papers, they show that this translational regulation also involves a purine-rich R-motif. They also observed that in the absence of pathogens, many defense-related genes show a suppression of translation that is alleviated upon pathogen exposure, indicating that a global translational reprogramming occurs. In the second new paper (10.1038/nature22372), the authors engineered translational regulation into a constitutively active immune receptor gene, snc1 or the defense regulator NPR1. In the absence of translational repression, snc1 and NPR1 expression confer broad-spectrum resistance, but with a substantial growth penalty. When expression is controlled by the repressive uORFs and R-motif, plants carrying these constructs show enhanced disease resistance without an accompanying growth penalty. Nature. Tags: Biotic Interactions, Molecular Biology, Signals and Responses
Plants fight back when pathogens attack, and in the first of a pair of papers Xu et al. have revealed a new insight into this response by demonstrating a significant translational upregulation of many defense-response mRNAs (10.1038/nature22371). Previously, this group showed that the translation of a key regulator of transcriptional responses to pathogens, TBF1, is itself regulated by the presence of two upstream open reading frames (uORFs). In the first of the new papers, they show that this translational regulation also involves a purine-rich R-motif. They also observed that in the absence of pathogens, many defense-related genes show a suppression of translation that is alleviated upon pathogen exposure, indicating that a global translational reprogramming occurs. In the second new paper (10.1038/nature22372), the authors engineered translational regulation into a constitutively active immune receptor gene, snc1 or the defense regulator NPR1. In the absence of translational repression, snc1 and NPR1 expression confer broad-spectrum resistance, but with a substantial growth penalty. When expression is controlled by the repressive uORFs and R-motif, plants carrying these constructs show enhanced disease resistance without an accompanying growth penalty. Nature. Tags: Biotic Interactions, Molecular Biology, Signals and Responses
Widespread biological response to rapid warming on the Antarctic Peninsula
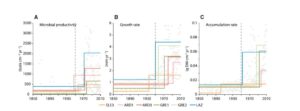 Amesbury et al. examined stratified cores from moss banks across the Antarctic Peninsula (AP) as a paleoclimate archive. They measured moss accumulation rate, growth rate, microbial productivity, and carbon-isotope discrimination (Δ13C, as a proxy for photosynthetic assimilation rate). Their data reveal a “widespread biological response to recent rapid warming” especially since about 1950, with spatial and temporal variability small compared to overall trends. The increase in Δ13C is consistent with warm, dry growing conditions. The authors conclude, “terrestrial plant communities and soils will undergo substantial alteration even with only modest further increases in temperature. These changes, combined with increased ice-free land areas from glacier retreat, will drive large-scale alteration to the biological functioning, appearance, and landscape of the AP over the rest of the 21st century and beyond.” Curr. Biol. 10.1016/j.cub.2017.04.034 Tags: Ecology
Amesbury et al. examined stratified cores from moss banks across the Antarctic Peninsula (AP) as a paleoclimate archive. They measured moss accumulation rate, growth rate, microbial productivity, and carbon-isotope discrimination (Δ13C, as a proxy for photosynthetic assimilation rate). Their data reveal a “widespread biological response to recent rapid warming” especially since about 1950, with spatial and temporal variability small compared to overall trends. The increase in Δ13C is consistent with warm, dry growing conditions. The authors conclude, “terrestrial plant communities and soils will undergo substantial alteration even with only modest further increases in temperature. These changes, combined with increased ice-free land areas from glacier retreat, will drive large-scale alteration to the biological functioning, appearance, and landscape of the AP over the rest of the 21st century and beyond.” Curr. Biol. 10.1016/j.cub.2017.04.034 Tags: Ecology




Leave a Reply
Want to join the discussion?Feel free to contribute!